|
Biodiversity Data Journal :
Data paper
|
|
Corresponding author:
Academic editor: Axel Hausmann
Received: 17 Sep 2014 | Accepted: 24 Feb 2015 | Published: 04 Mar 2015
© 2015 Rodolphe Rougerie, Carlos Lopez-Vaamonde, Thomas Barnouin, Julien Delnatte, Nicolas Moulin, Thierry Noblecourt, Benoît Nusillard, Guillem Parmain, Fabien Soldati, Christophe Bouget
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Rougerie R, Lopez-Vaamonde C, Barnouin T, Delnatte J, Moulin N, Noblecourt T, Nusillard B, Parmain G, Soldati F, Bouget C (2015) PASSIFOR: A reference library of DNA barcodes for French saproxylic beetles (Insecta, Coleoptera). Biodiversity Data Journal 3: e4078. https://doi.org/10.3897/BDJ.3.e4078
|
|
Abstract
Saproxylic beetles – associated with dead wood or with other insects, fungi and microorganisms that decompose it – play a major role in forest nutrient cycling. They are important ecosystem service providers and are used as key bio-indicators of old-growth forests. In France alone, where the present study took place, there are about 2500 species distributed within 71 families. This high diversity represents a major challenge for specimen sorting and identification.
The PASSIFOR project aims at developing a DNA metabarcoding approach to facilitate and enhance the monitoring of saproxylic beetles as indicators in ecological studies. As a first step toward that goal we assembled a library of DNA barcodes using the standard genetic marker for animals, i.e. a portion of the COI mitochondrial gene. In the present contribution, we release a library including 656 records representing 410 species in 40 different families. Species were identified by expert taxonomists, and each record is linked to a voucher specimen to enable future morphological examination. We also highlight and briefly discuss cases of low interspecific divergences, as well as cases of high intraspecific divergences that might represent cases of overlooked or cryptic diversity.
Keywords
DNA barcoding, COI, molecular identification, cryptic diversity, Coleoptera, forest insects, ecological indicators.
Introduction
Forests ecosystems cover nearly 30% of the total land surface globally and host most of the terrestrial biodiversity. They are highly complex systems whose functioning and sustainability depends on a range of spatially and temporally dynamic abiotic and biotic factors. To monitor or diagnose forest ecosystems, ecologists have historically used both physico-chemical and biological indicators. Among the latters, saproxylic beetles – associated with dead wood or with other insects, fungi and microorganisms that decompose it – have been used as key bio-indicators of old-growth forests in both temperate and boreal regions of the globe (but see
In conservation biology studies, saproxylic beetles have often been studied through the perspective of focal species (often endangered/patrimonial species (e.g.
DNA-based identification, and the development of metagenomic approaches using Next Generation Sequencing (NGS) technologies, hold strong promise to overcome this impediment and may alleviate funding and time constraints for large-scale studies on these insects. Molecular identification of species has seen a considerable and rapid development over the past decade following the introduction of DNA barcoding by
The PASSIFOR project, initiated by the National Research Institute of Science and Technology for Environment and Agriculture (IRSTEA) and by the National Institute for Agricultural Research (INRA), aims at developing a DNA metabarcoding approach for French species of saproxylic beetles to facilitate and enhance the use of these insects as forest indicators. As a first step toward that goal, we present and release in this paper a DNA barcode reference library for these insects, including 656 records representing 410 species in 251 genera and 40 different families. This library represents about 16% of the national fauna and we expect that its development in the next future will further contribute to the assembly of a DNA barcode library for European beetles. Remarkable progress was recently accomplished toward that goal with the published results of national campaigns in Finland (
General description
This library aims to provide an authoritative reference library for the DNA-based species identification of French saproxylic beetles, in order to facilitate the use of DNA metabarcoding in biodiversity monitoring networks focusing on these forest insects. It is also expected to develop the use of DNA barcodes by the community of coleopterists, in combination with characters from the morphology, ecology and biogeography of species, to address taxonomic questions.
Because the available funding for this project was too limited to develop an exhaustive library for French saproxylic beetles (ca. 2500 species) or to allow the documentation of intraspecific and geographical patterns of genetic variation, our initial objective has been to target a broad taxonomic coverage, favoring taxonomic diversity at the family, genus and species levels. Only a few species complex or notoriously difficult genera (e.g. Ampedus in family Elateridae) were more densely sampled.
The PASSIFOR library uses the standard DNA barcode for animals, i.e. a 658bp fragment of the COI mitochondrial gene.
Species identifications were provided by expert taxonomists for these groups. All records were initially identified on the basis of morphological examination, and voucher specimens are preserved in the collections of the taxonomists as references for these records. Any future change in the taxonomy/nomenclature of these insects will be reported in the PASSIFOR library, after authoritative validation by the taxonomists.
Project description
PASSIFOR: stands for (in French) "Proposition d'Amélioration du Système de Suivi de la bIodiversité FORestière": Proposal toward improving monitoring of forest biodiversity.
PIs: Christophe Bouget (IRSTEA, Nogent-Sur-Vernisson) & Carlos Lopez-Vaamonde (INRA, Orléans)
Postdoctoral fellow: Rodolphe Rougerie (INRA, Orléans)
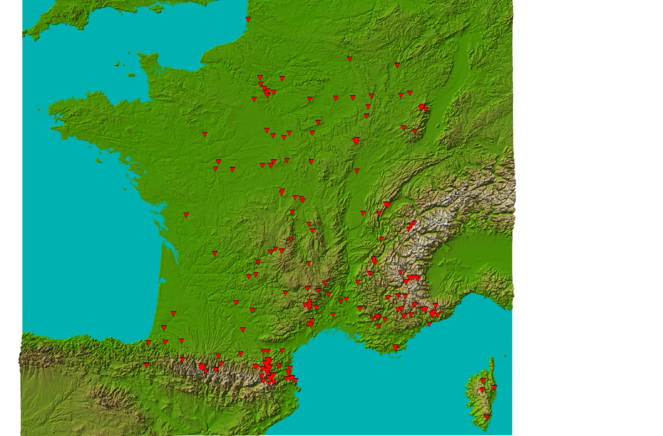
Western Europe: France (99.1% of the samples), Czech Republic, Italy, Spain, and Morocco.
This project is supported by a grant from the French Ministry of Agriculture (MAAF) to IRSTEA (CB) and INRA (CLV). Sequencing of DNA barcodes also benefitted from funding by Genome Canada and the Ontario Genomics Institute (OGI) to the International Barcode of Life Project (iBOL).
Sampling methods
The PASSIFOR library focuses on French species within 40 different families of saproxylic beetles.
Tissue samples for DNA extraction were collected mostly from dry collection specimens; only a limited number of samples were preserved in 95%-ethanol. All specimens were photographed and specimen data were compiled in excel spreadsheets for submission to BOLD.
Most specimens were sampled by RR and CLV in the National Laboratory for Forest Entomology, Quillan, France. GP, TB and BN assisted in sampling and in databasing records in their institutional collections. NM sampled specimens in his own reference collection, while JD selected and shipped a selection of specimens (Elateridae, especially members of genus Ampedus) to INRA Orleans where RR handled tissue sampling, photography and databasing.
All tissue samples were assembled in 96-well plates in which one well (location H12) was left empty to serve as a negative control. After sequencing and upload of sequences into BOLD, DNA barcodes were compared through classical analyses of genetic distances (blast hits, NJ trees) to conspecific records, when existing, in other accessible DNA barcoding projects/campaigns. Discordances between DNA results and taxonomy derived from morphology (DNA barcodes shared by distinct species, deep intra-specific splits (>2%)) led to re-examination of the specimens; collegial discussions were initiated to address these issues by revealing possible cases of mis-identification or cross-contamination.
The construction of the PASSIFOR library can be divided into two main steps:
- Specimen sampling and data compilation:
- tissue sampling. Using flame-decontaminated forceps, we usually pulled one leg from each specimen sampled. Occasionally, when these appendages were difficult to reach, we used the antenna or hindwing of the insect. For the smallest species, we sometimes used up to three of these body parts (usually several legs). Only in one case, a tiny representative of the Scolytinae Ernoporicus fagi, did we use the whole insect and as a consequence did not preserve any voucher specimen.
- photography. Each specimen was photographed individually along with a scale.
- data compilation. We used standard BOLD spreadsheets to compile:
- voucher information: SampleID (a unique BOLD identifier for the specimen; also added on a label pinned with the voucher specimen) and institution storing.
- Taxonomy data: higher level taxonomy; species identification; identifier, including contact information.
- Specimen details: sex (when available); reproduction mode; life stage; type of tissue used (for most specimens); collecting method (when available).
- Collection data: collectors; date collected; country; administrative region and department; sector; exact site; latitude, longitude and elevation (when available).
- upload to BOLD. Following the standard BOLD procedure for DNA barcode library construction, we created a dedicated project in BOLD. This project (code PSFOR, publicly accessible) hosts records for all the samples processed (including failures), whereas the actual PASSIFOR library (dataset DS-PSFOR01, see the Data resources section below) only includes records successfully sequenced and subsequently validated by taxonomists.
- Sequencing of DNA barcodes: The Canadian Centre for DNA Barcoding (CCDB), hosted by the Biodiversity Institute of Ontario (BIO) at the University of Guelph, Ontario, Canada) processed the tissue samples; all operations were carried out following the standard high-throughput protocols in place at CCDB and available from http://ccdb.ca/resources.php. For PCR amplification, we used a primer cocktail combining the LCO1490/HCO2198 pair (
Folmer et al. 1994 ) with the LepF1/LepR1 pair (Hebert et al. 2004 ) for amplification of the full-length (658bp) DNA barcode region of the COI gene. Samples failing to amplify with these primers were alternatively processed using internal primers targeting shorter fragment; MLepR2 (Hebert et al. 2013 ) was used along with LCO1490/LepF1, and MLepF1 (Hajibabaei et al. 2006 ) was used with HCO1498/LepR1 to target fragments of 307bp and 407bp, respectively. Unpurified PCR fragments were sequenced in both directions using an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). CodonCode (CodonCode Corporation, Centerville, MA) was used for trimming primers, contig assembly and sequence editing; alignment was straightforward in absence of indels and the sequences, along with corresponding trace files, were uploaded to BOLD.
Geographic coverage
41.7 and 50.5 Latitude; -1.6 and 9.5 Longitude.
Taxonomic coverage
The PASSIFOR library comprises 656 records for saproxylic beetles belonging to 40 different families. They represent 410 species in 251 genera. Table
Taxonomic coverage of the PASSIFOR library giving details of the number of records, genera and species sampled within each of the 40 families included (ordered alphabetically).
| Family | Records | Genera | Species |
|---|---|---|---|
| Anthribidae | 11 | 9 | 9 |
| Biphyllidae | 2 | 1 | 1 |
| Bostrichidae | 1 | 1 | 1 |
| Brentidae | 1 | 1 | 1 |
| Buprestidae | 7 | 6 | 7 |
| Cerambycidae | 165 | 69 | 115 |
| Cerophytidae | 1 | 1 | 1 |
| Ciidae | 1 | 1 | 1 |
| Cleridae | 12 | 5 | 6 |
| Curculionidae | 54 | 19 | 31 |
| Elateridae | 151 | 22 | 57 |
| Endomychidae | 2 | 1 | 1 |
| Erotylidae | 2 | 2 | 2 |
| Eucinetidae | 1 | 1 | 1 |
| Eucnemidae | 8 | 4 | 5 |
| Histeridae | 1 | 1 | 1 |
| Laemophloeidae | 2 | 2 | 2 |
| Leiodidae | 3 | 2 | 2 |
| Lucanidae | 8 | 4 | 4 |
| Lycidae | 6 | 5 | 5 |
| Lymexylidae | 4 | 2 | 2 |
| Melandryidae | 24 | 12 | 16 |
| Monotomidae | 9 | 1 | 7 |
| Mycetophagidae | 15 | 4 | 11 |
| Nitidulidae | 7 | 3 | 6 |
| Nosodendridae | 1 | 1 | 1 |
| Oedemeridae | 11 | 4 | 8 |
| Prostomidae | 1 | 1 | 1 |
| Ptinidae | 30 | 12 | 23 |
| Pyrochroidae | 5 | 2 | 3 |
| Pythidae | 1 | 1 | 1 |
| Salpingidae | 14 | 6 | 11 |
| Scarabaeidae | 16 | 5 | 14 |
| Silvanidae | 2 | 2 | 2 |
| Sphindidae | 2 | 2 | 2 |
| Tenebrionidae | 51 | 21 | 32 |
| Tetratomidae | 1 | 1 | 1 |
| Trogidae | 2 | 1 | 1 |
| Trogossitidae | 14 | 8 | 9 |
| Zopheridae | 7 | 5 | 6 |
| Total | 656 | 251 | 410 |
The nomenclature used generally follows that in the eight volumes of the Catalogue of Palaearctic Coleoptera series, edited by Löbl and Smetana (see f.i
Usage rights
Data resources
The PASSIFOR library dataset can be downloaded from the Public Data Portal of BOLD in different formats (data as xml or tsv files, sequences and trace files as fasta and ab1 files). Alternatively, BOLD users can login and access the dataset via the Workbench platform of BOLD (see the public dataset list in the User Console page, under the name of first author); all records are also searchable within BOLD using the search function of the database.
The version of the library at the time of writing of this manuscript is also included as Suppl. materials
| Column label | Column description |
|---|---|
| processid | Unique identifier for the DNA sample. |
| sampleid | Unique identifier for the specimen and by extension the tissue sample used for DNA analysis. |
| recordID | Entry number in the database. |
| catalognum | Identifier for specimen assigned by formal collection upon accessioning. |
| fieldnum | Identifier for specimen assigned in the field. |
| institution_storing | The full name of the institution that has physical possession of the voucher specimen. |
| bin_uri | URI (Unique Resource Identifier) for the Barcode Index Number (BIN) to which the record belongs. |
| phylum_taxID | Taxonomic identifier of level Phylum |
| phylum_name | Phylum name |
| class_taxID | Taxonomic identifier of level Class |
| class_name | Class name |
| order_taxID | Taxonomic identifier of level Order |
| order_name | Order name |
| family_taxID | Taxonomic identifier of level Family |
| family_name | Family name |
| subfamily_taxID | Taxonomic identifier of level Subfamily |
| subfamily_name | Subfamily name |
| genus_taxID | Taxonomic identifier of level Genus |
| genus_name | Genus name |
| species_taxID | Taxonomic identifier of level Species |
| species_name | Species name |
| identification_provided_by | Full name of primary individual who assigned the specimen to a taxonomic group. |
| voucher_type | Status of the specimen in an accessioning process. |
| tissue_type | A brief description of the type of tissue or material analyzed. |
| collectors | The full or abbreviated names of the individuals or team responsible for collecting the sample in the field. |
| collectiondate | The date during which the sample was collected. |
| collectiondate_accuracy | A numerical representation of the precision of the Collection Date given in days and is represented as +/- the value. |
| lifestage | The age class or life stage of the specimen at the time of sampling. |
| sex | The sex of the specimen. |
| reproduction | The presumed method of reproduction. |
| extrainfo | A brief note or project term associated with the specimen for rapid analysis. |
| notes | General notes regarding the specimen. |
| lat | The geographic latitude (in decimal degrees) of the geographic center of a location. |
| lon | The geographic longitude (in decimal degrees) of the geographic center of a location. |
| coord_source | The source of the latitude and longitude measurements. |
| coord_accuracy | A decimal representation of the precision of the coordinates given in the decimalLatitude and decimalLongitude. |
| elev | Elevation of sampling site. Measured in meters relative to sea level. Negative values indicate a position below sea level. |
| depth | For organisms collected beneath the surface of a water body. Measured in meters below surface of water. |
| elev_accuracy | A numerical representation of the precision of the elevation given in meters and is represented as +/- the elevation value. |
| depth_accuracy | A numerical representation of the precision of the depth given in meters and is represented as +/- the depth value. |
| country | The full, unabbreviated name of the country, major political unit, or ocean in which the organism was collected. |
| province | The full, unabbreviated name of the state, province, territory, or prefecture (i.e., the next smallest political region below Country) in which the organism was collected. |
| region | The full, unabbreviated name of the county, shire, municipality, or park (i.e., the next smallest political region below province/state) in which the organism was collected. |
| sector | The full, unabbreviated name of the lake, conservation area or sector of park in which the organism was collected. |
| exactsite | Additional text descriptions regarding the exact location of the collection site relative to a geographic or biologically relevant landmark. |
Additional information
In the following sections we provide a quick description of the results of DNA barcode analyses as carried out using the analytical tools available through the BOLD's workbench at the time of writing of this manuscript.
Sequence composition
The summary statistics for nucleotide frequency distribution are provided in Table
Nucleotide frequency distribution for sequences (>400bp, 597 sequences analyzed) in the PASSIFOR library.
| Min | Mean | Max | SE | |
|---|---|---|---|---|
| G % | 13.37 | 16.17 | 21.73 | 0.04 |
| C % | 12.61 | 19.82 | 27.68 | 0.13 |
| A % | 25.31 | 29.74 | 34.15 | 0.06 |
| T % | 26.44 | 34.26 | 44.07 | 0.15 |
| GC % | 25.99 | 35.99 | 46.81 | 0.14 |
| GC % Codon Pos 1 | 34.65 | 46.84 | 54.72 | 0.13 |
| GC % Codon Pos 2 | 38.3 | 42.65 | 46.44 | 0.04 |
| GC % Codon Pos 3 | 1.94 | 18.43 | 43.77 | 0.31 |
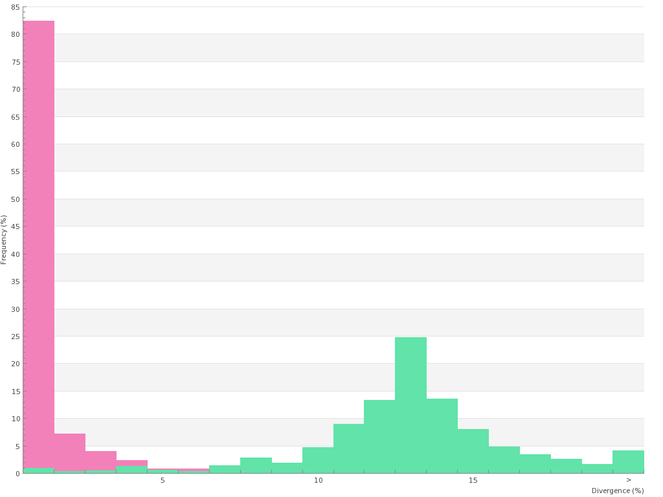
Analyses of genetic distances
All sequence analyses were carried out in BOLD using Kimura-2 parameters (K2P) distances with BOLD handling the sequence alignment. Alternative alignment methods were tested (including the use of sequences aligned "as uploaded") and proved to have no impact on the results.
All 656 sequences of the library where used to build a Neighbor-Joining (NJ) tree as illustrated in Suppl. material
Summary of distance (K2P) variations at species, genus and family levels, as calculated with BOLD from 597 records of the PASSIFOR library with DNA barcodes longer than 400bp.
| n | Taxa | Comparisons | Min Dist(%) | Mean Dist(%) | Max Dist(%) | SE Dist(%) | |
|---|---|---|---|---|---|---|---|
| Within Species | 334 | 125 | 458 | 0 | 0.85 | 14.93 | 0 |
| Within Genus | 362 | 73 | 3152 | 0 | 12.5 | 27.14 | 0 |
| Within Family | 573 | 25 | 20617 | 9.54 | 21.9 | 39.13 | 0 |
Discrepancies between current taxonomy and DNA barcode results
While we are aware of the limitation of our dataset to address taxonomic questions in cases where DNA barcodes and current taxonomy reveal a possible discordance, we report here two categories of apparent conflicts between the results from DNA barcode analyses and species identifications derived from morphology.
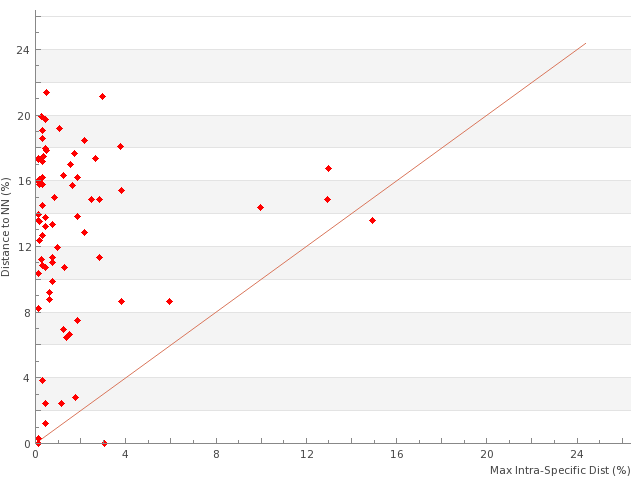
- High intraspecific divergence (>2%) were observed in 7 species (Table
4 ). All these cases require further sampling and investigation to figure if they represent cases of overlooked or cryptic diversity, or if they may represent geographical population structure, ancestral polymorphisms, or variation resulting from Wolbachia infections (Smith et al. 2012 ). As an example, in the Tenebrionidae Nalassus ecoffeti, where intraspecific genetic distance is as high as 13.2%, our results suggest the possible validity of the currently synonymized Pyrenean species N. temperei Ardoin, 1958 (F. Soldati, personal communication). - Low interspecific divergences (<2%) were observed in 6 pairs of species, 1 triplet, and 2 pairs of subspecies (Table
5 ). In total, of the 410 species sampled in the PASSIFOR library, 15 (3.6%) fall in this category of low to null interspecific distances. Here again, these cases will require additional sampling and further investigation to understand if our results reflect cases of overlooked synonymy (as may be the case in the pairs Ampedus pomorum / A. nemoralis, Anastrangalia dubia / A. reyi (the second originally described as a mere variety of the former)), introgression through past or ongoing hybridization, or recent speciation resulting in low level of divergence (e.g. in the pairs Pityophagus ferrugineus / P. laevior and Ampedus pomonae / A. sanguinolentus). In fact, our results only revealed two cases of strictly shared DNA barcodes (one pair and one triplet within the taxonomically difficult genus Ampedus), although results for Central European samples of Anastrangalia dubia and A. reyi (Hendrich et al. 2014 ) confirmed that the two species cannot be distinguished using their DNA barcodes.
List of species within the PASSIFOR library (sequence length>400 bp; 597 records, 388 species) with more than 2% intraspecific divergence (N = number of records).
| Family | Species | N | Max. Intrasp. (%) |
|---|---|---|---|
| Cerambycidae | Alosterna tabacicolor | 3 | 11.2 |
| Cerambycidae | Tetrops praeustus | 2 | 11.8 |
| Cleridae | Thanasimus formicarius | 2 | 11.5 |
| Cleridae | Tillus elongatus | 4 | 8.8 |
| Elateridae | Melanotus castanipes | 2 | 5.7 |
| Elateridae | Melanotus villosus | 3 | 4.5 |
| Tenebrionidae | Nalassus ecoffeti | 5 | 13.2 |
List of species and subspecies pairs/triplet within the PASSIFOR library for which the minimum distance to the nearest heterospecific or heterosubspecific record is below 2% (number of records for each taxon is given within brackets next to its name).
| Family | Species pairs & triplet | Min. intersp. (%) |
| Cerambycidae | Anastrangalia dubia (3) / A. reyi (1) | 0.47 |
| Cerambycidae | Chlorophorus ruficornis (1) / C. sartor (1) | 1.1 |
| Cerambycidae | Paracorymbia hybrida (1) / P. maculicornis (1) | 0.92 |
| Elateridae | Ampedus cardinalis (3) / A. praestus (2) / A. melonii (1) | 0 |
| Elateridae | Ampedus pomonae (1) / A. sanguinolentus (1) | 1.61 |
| Elateridae | Ampedus pomorum (9) / A. nemoralis (3) | 0 |
| Lucanidae | Lucanus cervus (1) / L. cervus fabiani (1) | 0 |
| Nitidulidae | Pityophagus ferrugineus (1) / P. laevior (1) | 1.88 |
| Scarabaeidae | Protaetia cuprea (1) / P. cuprea metallica (1) | 1.22 |
Acknowledgements
We are thankful to Benoît Dodelin (Lyon, France) for providing additional material in genus Ampedus, and to Hervé Brustel and Lionel Valladares of the PURPAN Engineering School (Toulouse, France) for their discussions of taxonomic questions.
We also gratefully acknowledge the support from colleagues at the Canadian Centre for DNA Barcoding (University of Guelph, Ontario, Canada), and in particular Natalia Ivanova, Jayme Sones and Evgeny Zakharov who carefully and efficiently supervised the processing of the material analyzed in the present work.
Author contributions
Contributed to:
- Study design: CB, CLV, RR
- Specimen sampling, databasing: JD, CLV, NM, BN, GP, RR
- Sequence analyses: RR
- Taxonomic expertise, result validation: TB, CB, JD, NM, TN, BN, GP, FS
- Writing of manuscript: RR
- Editing/comments to the manuscript: TB, CB, JD, CLV, NM, TN, BN, GP, RR, FS
References
- Evaluation of window flight traps and ethanol lures for effectiveness at monitoring dead wood associated beetles.Agricultural and Forest Entomology11:143‑152.
- Key features for saproxylic biodiversity from rapid habitat assessment in temperate forests.Ecological Indicators36:656‑664.
- Modelling habitat and spatial distribution of an endangered longhorn beetle – A case study for saproxylic insect conservation.Biological Conservation137(3):372‑381. https://doi.org/10.1016/j.biocon.2007.02.025
- Saproxylic beetle assemblages in the Mediterranean region: Impact of forest management on richness and structure.Forest Ecology and Management259(8):1376‑1384. https://doi.org/10.1016/j.foreco.2010.01.004
- Diagnosing Mitochondrial DNA Diversity: Applications of a Sentinel Gene Approach.Journal of Molecular Evolution66(4):362‑367. https://doi.org/10.1007/s00239-008-9088-2
- From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity.Trends in Ecology & Evolutionin press:1‑6. https://doi.org/10.1016/j.tree.2014.08.001
- DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates.Molecular Marine Biology and Biotechnology3:294‑299.
- Saproxylic insect ecology and the sustainable management of forests.Annual Review of Ecology and Systematics33:1‑23.
- The conservation of saproxylic insects in tropical forests: a research agenda.Journal of Insect Conservation3:67‑74.
- DNA barcodes distinguish species of tropical Lepidoptera.Proceedings of the National Academy of Sciences103(4):968‑971.
- Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos.PLoS One6(4):e17497. [Ineng]. https://doi.org/10.1371/journal.pone.0017497
- Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences270:313‑321.
- Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator.Proceedings of the National Academy of Sciences101(41):14812‑14817. https://doi.org/10.1073/pnas.0406166101
- A DNA 'barcode blitz': rapid digitization and sequencing of a natural history collection.PLoS One8(7):e68535. [Ineng]. https://doi.org/10.1371/journal.pone.0068535
- A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD.Molecular Ecology Resourcesn/a:n/a‑n/a. https://doi.org/10.1111/1755-0998.12354
- The effects of forest age on saproxylic beetle biodiversity: implications of shortened and extended rotation lengths in a French oak high forest.Insect Conservation and Diversity6(3):396‑410. https://doi.org/10.1111/j.1752-4598.2012.00214.x
- Catalogue of Palaearctic Coleoptera Volume I: Archostemata-Myxophaga-Adephaga.Apollo Books,Stenstrup, Denmark,819pp.
- Barcoding Beetles: A Regional Survey of 1872 Species Reveals High Identification Success and Unusually Deep Interspecific Divergences.PLoS ONE9(9):e108651. https://doi.org/10.1371/journal.pone.0108651
- Breaking down complex Saproxylic communities: understanding sub-networks structure and implications to network robustness.PLoS One7(9):e45062. [Ineng]. https://doi.org/10.1371/journal.pone.0045062
- BOLD: The Barcode of Life Data System (http://www.barcodinglife.org).Molecular Ecology Notes7(3):355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- Next-generation sequencing technologies for environmental DNA research.Mol Ecol21(8):1794‑805. [Ineng]. https://doi.org/10.1111/j.1365-294X.2012.05538.x
- Wolbachia and DNA Barcoding Insects: Patterns, Potential, and Problems.PLoS One7(5):e36514. https://doi.org/10.1371/journal.pone.0036514
- Towards next-generation biodiversity assessment using DNA metabarcoding.Molecular Ecology21:2045‑2050.
- Next generation molecular ecology.Mol Ecol19 Suppl 1:1‑3. [Ineng]. https://doi.org/10.1111/j.1365-294X.2009.04489.x
- Catalogue des Coléoptères de France.Association Roussillonnaise d'Entomologie,Perpignan, France,1056pp.
- Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring.Methods in Ecology and Evolution3(4):613‑623. https://doi.org/10.1111/j.2041-210X.2012.00198.x
Supplementary materials
This excel spreadsheet includes information about all records in BOLD for the PASSIFOR library at the time of writing. It contains specimen data and sequence information, including GenBank accession numbers.
Sequences in fasta format for the fragment of the COI mtDNA gene used as a standard DNA barcode in animals. Each sequence is identified by a chain of characters consisting of, in the following order and separated by pipes: processID, sampleID, species_name, DNA marker
NJ tree resulting from the analysis with BOLD of the 656 DNA barcode sequences of the PASSIFOR library. Parameters for tree reconstruction are as follow: distance model: K2P; alignment method: BOLD aligner; sequence length: >200 bp; pairwise deletion option; all three codon positions included.
This table lists K2P distances for all pairwise comparisons between conspecific records in the PASSIFOR library (only DNA barcodes longer than 400bp); distances are calculated in BOLD (www.boldsystems.org).
For the PASSIFOR library (only DNA barcodes longer than 400bp), this table lists K2P distances for all pairwise comparisons between heterospecific records of the same genus; distances are calculated in BOLD (www.boldsystems.org).
This table provides, for each species of the PASSIFOR library with sequences longer than 400bp, mean and maximum intraspecific distances (non-applicable (N/A) for species represented as singletons in our dataset) as well as the distance to nearest neighbor (NN) within the library and its identification.