|
Biodiversity Data Journal :
Software description
|
|
Corresponding author:
Academic editor: Scott Chamberlain
Received: 25 May 2015 | Accepted: 08 Jul 2015 | Published: 28 Jul 2015
© 2015 Alejandro Ruete
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Ruete A (2015) Displaying bias in sampling effort of data accessed from biodiversity databases using ignorance maps. Biodiversity Data Journal 3: e5361. https://doi.org/10.3897/BDJ.3.e5361
|
|
Abstract
Background
Open-access biodiversity databases including mainly citizen science data make temporally and spatially extensive species’ observation data available to a wide range of users. Such data have limitations however, which include: sampling bias in favour of recorder distribution, lack of survey effort assessment, and lack of coverage of the distribution of all organisms. These limitations are not always recorded, while any technical assessment or scientific research based on such data should include an evaluation of the uncertainty of its source data and researchers should acknowledge this information in their analysis. The here proposed maps of ignorance are a critical and easy way to implement a tool to not only visually explore the quality of the data, but also to filter out unreliable results.
New information
I present simple algorithms to display ignorance maps as a tool to report the spatial distribution of the bias and lack of sampling effort across a study region. Ignorance scores are expressed solely based on raw data in order to rely on the fewest assumptions possible. Therefore there is no prediction or estimation involved. The rationale is based on the assumption that it is appropriate to use species groups as a surrogate for sampling effort because it is likely that an entire group of species observed by similar methods will share similar bias. Simple algorithms are then used to transform raw data into ignorance scores scaled 0-1 that are easily comparable and scalable. Because of the need to perform calculations over big datasets, simplicity is crucial for web-based implementations on infrastructures for biodiversity information.
With these algorithms, any infrastructure for biodiversity information can offer a quality report of the observations accessed through them. Users can specify a reference taxonomic group and a time frame according to the research question. The potential of this tool lies in the simplicity of its algorithms and in the lack of assumptions made about the bias distribution, giving the user the freedom to tailor analyses to their specific needs.
Keywords
Biodiversity database, citizen-science data, presence-only data, sampling effort, spatial bias, species distribution model, Swedish Lifewatch
Introduction
“The greatest enemy of knowledge is not ignorance; it is the illusion of knowledge.” Daniel J. Boorstin
The emergence of open-access databases on diverse kinds of environmental data (e.g. www.worldclim.org; www.climond.org) and species occurrences data (e.g. www.gbif.org) has led to a rapid increase in biogeographical studies developing new theories, methodologies and applications for nature conservancy (
For most species, raw distributional data accessible in biodiversity databases are presence data coming from museums, herbaria, inventories, or citizen science programs, and are the result of a vast number of observers collecting data over a large time span with no specific sampling design (
All these issues stemming from the quality of the raw data can be ameliorated by the use of parallel “maps of ignorance” to provide information on sampling coverage and reliability (
I present simple algorithms to create and display ignorance maps based upon presence-only observation records. The algorithms are thought to be general enough to be implemented as web-based tools to download ignorance scores in the form of raster images. Ignorance maps will serve to properly inform users of the bias inherent to the data and to provide them with tools to properly analyse the raw data provided. The approach presented here is in line with the need identified by
Project description
Ignorance maps of raw data accessed from species observation databases
Worldwide; example data from Sweden
Rationale and assumptions
The aim is to provide ignorance maps that are easily comparable and easily scalable, to report the spatial distribution of sampling effort (or lack of it). Therefore the obvious choice is to represent ignorance on a scale of 0 to 1 (1 being absolute ignorance and 0 being absolute certainty or credibility in the data). There are several approaches to incorporate sampling effort to different analysis of richness, species distributions and trends in population abundance (
Observations are reported by people with varied field skills and accuracy. Because of the intrinsic characteristics of the reports (e.g. voluntary, non-systematic), biodiversity datasets have a considerable spatial and temporal bias. However, observers are assumed to be fond of or specialist on one or more taxonomic groups (e.g. family, order), rather than on individual species. Since it is likely that an entire group of species observed by similar methods (henceforth a reference taxonomic group) will share similar bias (
There are some considerations to take into account before describing the algorithms. First, the reference target group should only include species that are assumed to be sampled with the same methodology, to keep the sampling bias consistent (
Algorithms overview
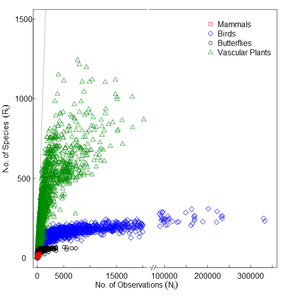
The sampling behaviour that characterizes observers differs among reference taxonomic groups. For some groups like vascular plants or bryophytes observers typically inventory confined areas (sites) reporting every species they observe, aiming to cover as many sites as possible. In these cases, raw observation counts per grid cell i (Ni) better represent the sampling intensity and species discovery (Fig.
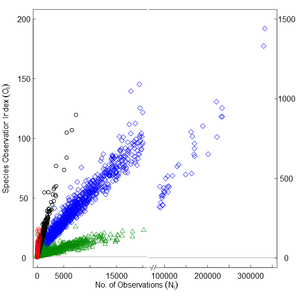
a) Species discovery plot (Ni vs Ri) and b) species observation index (Oi) as a function of the number of observations per grid cell (Ni; Suppl. materials
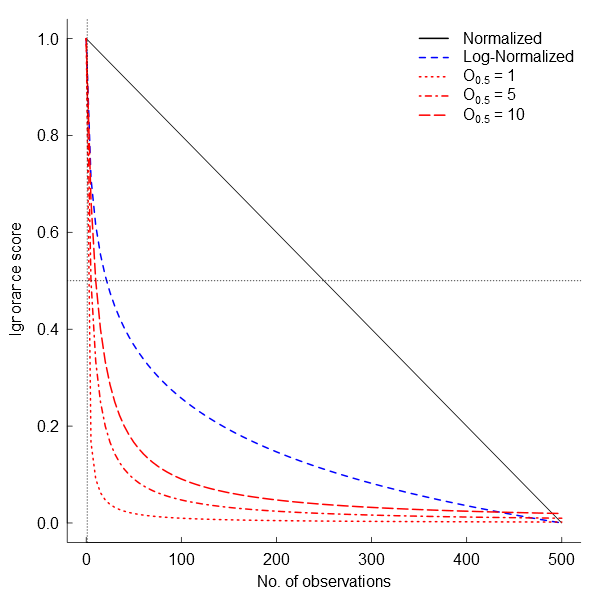
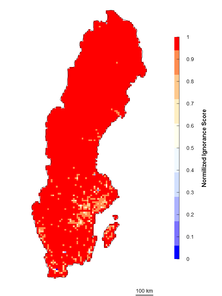
The first and easiest way to transform observation counts into a 0-1 scale of ignorance (I) is by using normalized data (henceforth the Normalization approach): \(I_{i}=1-N_{i}/N_{m}\) where Nm is the maximum number of observations per grid cell of the dataset. Then 0 represents the maximum certainty of the data corresponding to the maximum number of observations recorded in the entire dataset (Fig.
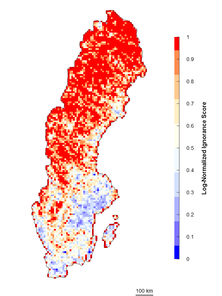
In many cases there are sites that are more than sufficiently sampled (i.e. long right tails in the probability distribution of observations) but the relative influence of these sites on our certainty may not be linear. In these cases, when it is relevant to separate sites with “few” observations from sites with “enough” observations, logarithmic transformations are preferred (Fig.
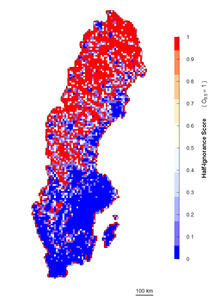
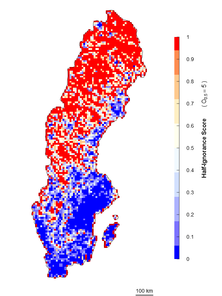
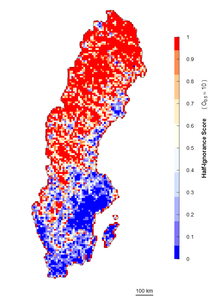
An alternative approach is an algorithm independent of the maximum number of observations. It estimates ignorance scores making data relative to a reference number of observations that is considered to be enough to reduce the ignorance score by half (henceforth the Half-ignorance approach). In this case, ignorance scores are defined as \(I_{i} = O_{0.5}/(N_{i}+O_{0.5})\) (Fig.
Raw observations (a; Suppl. material
It is important to highlight that the size of the grid cells (i.e. resolution) will affect the results of all implemented algorithms. For example, consider the simple case where one large grid cell is made up of four smaller cells of which three cells are empty and only one cell scores all the reported observations. In this case the spatial distribution of recording effort will look very different when mapped at a high or low resolution. Sensitivity to spatial resolution is a common problem on studies summarizing biodiversity data on arbitrary grid cells, and the relevance of this problem has to be evaluated for each study in light of the question or hypothesis tested (
This project was framed within and funded by the Swedish LifeWatch.
Web location (URIs)
Technical specification
Implementation
Implements specification
The code provided in the repository is implemented as an HTML application with a local R server through the package shiny. The core algorithms introduced here are not dependent on any language and can be used independently or be implemented on biodiversity data portals. For example, the Swedish Lifewatch analysis portal is currently implementing this algorithms in the JAVA language.
The R code provided is adapted to run under the shiny server framework, however those who need can find the core algorithms in plain R language in the file "SLWApp/server.r" provided in the repository. This R code and examples will remain in the repository for individual implementations and modifications.
In order to use the R code as is with other species the requirements are:
- a raster image where each pixel summarizes the total number of observations recorded for the reference taxonomic group during the desired time frame
- a raster image where each pixel summarizes the total number of individual species within the reference taxonomic group observed during the desired time frame
- a raster image where each pixel summarizes the total number of observations recorded for the focal species during the desired time frame
- (Optional) a shape file (.shp) with the contour of the study region
Note that all raster images must have the exact same extent and resolution. In the examples presented here these raster images were created transforming the grid-based summary tables obtained from the Swedish LifeWatch analysis portal into. tiff georeferenced images. Some portals (e.g. GBIF) will only be able to download individual observation data points, in which case the user will need to summarize the data into raster images.
Note as well that although this code is implemented to calculate ignorance scores per pixel, the algorithms can be applied to summaries of irregular areas.
Audience
Database users can assess, with three alternative algorithms, the spatial bias of the sampling effort and relative amount of knowledge gained for any reference taxonomic group, and download these mapped ignorance data as GIS-layers. End-users will be able to individually set the scale, resolution, time frame, and reference taxonomic groups of interest to assess the utility of the observations reported in the database. Potential target users of the ignorance maps are: 1) consultants performing environmental impact assessments (e.g. they could use ignorance maps to make precautionary statements about lack of knowledge about species of special conservation interest on areas where projects are intended to be developed); 2) observers (e.g. they might be interested in locating under-sampled areas to be targeted on their next campaign); and 3) researchers (they might benefit in many different ways, some of which we describe below).
The most obvious use for ignorance maps is to mask out from other raster layers derived from the raw data (e.g. estimates of pseudo-absence or population abundance) areas of high uncertainty, excluding them from further analyses. A user-defined ignorance threshold could be used to generate pseudo-absences on sites where focal species are likely to be absent given the species has not been observed and that the site counts with high sampling effort for the reference taxonomic group (
Ignorance maps are of particular interest for species distribution modelling (SDM), as estimates can be improved by incorporating information on how recording effort varies spatially (
Within the Bayesian framework SDMs could also benefit by using ignorance scores to inform a priori probability distributions (
Additional information
Conclusion
Dealing with uncertainty in presence-only citizen science data is necessary for a wide range of applications, and the development of an ignorance score as implemented here provides an appropriate scale to compare different taxa, and a straight forward and easily interpretable method of doing so. Any infrastructure for biodiversity information on virtually any web infrastructure can offer a quality report of the spatial bias of observations stored in databases implementing these simple algorithms. Quantifying recording effort in citizen science biodiversity datasets allows users to incorporate uncertainty into analyses of species’ richness and distributions, to identify unreliable analyses results, and to identify areas where further surveys are required. Users can specify a reference taxonomic group and a time frame according to the research question. The potential of this tool lies in the simplicity of its algorithms and the lack of assumptions made about the bias distribution, giving the user the freedom to tailor analyses to their specific needs.
Acknowledgements
I gratefully acknowledge the participation of colleagues from the Swedish Species Information Centre in discussions of preliminary versions of this implementation, particularly to Ulf Gärdenfors, Oskar Kindvall and Louise Mair.
References
- Prediction of potential areas of species distributions based on presence-only data.Environmental and Ecological Statistics12(1):27‑44. https://doi.org/10.1007/s10651-005-6816-2
- Evaluating protected area effectiveness using bird lists in the Australian Wet Tropics.Diversity and Distributions21(4):368‑378. https://doi.org/10.1111/ddi.12274
- The art of modelling range-shifting species.Methods in Ecology and Evolution1(4):330‑342. https://doi.org/10.1111/j.2041-210X.2010.00036.x
- Moving beyond static species distribution models in support of conservation biogeography.Diversity and Distributions16(3):321‑330. https://doi.org/10.1111/j.1472-4642.2010.00641.x
- Species distribution models in conservation biogeography: developments and challenges.Diversity and Distributions19(10):1217‑1223. https://doi.org/10.1111/ddi.12125
- Swedish LifeWatch ─ a biodiversity infrastructure integrating and reusing data from citizen science, monitoring and research.Human Computation1(2):1. https://doi.org/10.15346/hc.v1i2.6
- Field validation shows bias-corrected pseudo-absence selection is the best method for predictive species-distribution modelling.Diversity and Distributions20(12):1403‑1413. https://doi.org/10.1111/ddi.12249
- Local frequency as a key to interpreting species occurrence data when recording effort is not known.Methods in Ecology and Evolution3(1):195‑205. https://doi.org/10.1111/j.2041-210X.2011.00146.x
- Uncertainty and the measurement of terrestrial biodiversity gradients.Journal of Biogeography35(8):1335‑1336. https://doi.org/10.1111/j.1365-2699.2008.01955.x
- Limitations of biodiversity databases: case study on seed-plant diversity in tenerife, canary islands.Conservation Biology21(3):853‑863. https://doi.org/10.1111/j.1523-1739.2007.00686.x
- Species richness, hotspots, and the scale dependence of range maps in ecology and conservation.Proceedings of the National Academy of Sciences104(33):13384‑13389. https://doi.org/10.1073/pnas.0704469104
- The use of historical collections to estimate population trends: A case study using Swedish longhorn beetles (Coleoptera: Cerambycidae).Biological Conservation143(9):1940‑1950. https://doi.org/10.1016/j.biocon.2010.04.015
- Profiting from prior information in Bayesian analyses of ecological data.Journal of Applied Ecology42(6):1012‑1019. https://doi.org/10.1111/j.1365-2664.2005.01101.x
- Integrating fundamental concepts of ecology, biogeography, and sampling into effective ecological niche modeling and species distribution modeling.Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology146(4):789‑796. https://doi.org/10.1080/11263504.2012.740083
- Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data.Ecological Applications19(1):181‑197. https://doi.org/10.1890/07-2153.1
- Evaluation of museum collection data for use in biodiversity assessment.Conservation Biology15(3):648‑657. https://doi.org/10.1046/j.1523-1739.2001.015003648.x
- Correcting for variation in recording effort in analyses of diversity hotspots.Biodiversity Letters1(2):39. https://doi.org/10.2307/2999649
- Accounting for uncertainty when mapping species distributions: The need for maps of ignorance.Progress in Physical Geography35(2):211‑226. https://doi.org/10.1177/0309133311399491
- Analysing botanical collecting effort in Amazonia and correcting for it in species range estimation.Journal of Biogeography34(8):1388‑1399. https://doi.org/10.1111/j.1365-2699.2007.01716.x
- Evaluating citizen-based presence data for bird monitoring.Biological Conservation144(2):804‑810. https://doi.org/10.1016/j.biocon.2010.11.010
- Accounting for spatially biased sampling effort in presence-only species distribution modelling.Diversity and Distributions21(5):595‑608. https://doi.org/10.1111/ddi.12279
- The value of museum collections for research and society.BioScience54(1):66. https://doi.org/10.1641/0006-3568(2004)054[0066:TVOMCF]2.0.CO;2
- The effects of sampling bias and model complexity on the predictive performance of Maxent species distribution models.PLoS ONE8(2):e55158. https://doi.org/10.1371/journal.pone.0055158
Supplementary materials
The algorithms are designed to handle number of observations and number of species summarized per grid cells. Here I provide the. CSV files as downloaded from www.analysisportal.se. This is the format one is expected to get the summarized data for a biodiversity database. I also include data on the occurrence of two species (a common and a rare) for each reference taxonomic group.
The algorithms are designed to handle number of observations and number of species summarized per grid cells. Here I provide the raster images used for the examples provided in the R script available on the GitHUB repository. These images were produced from. CSV files downloaded from www.analysisportal.se
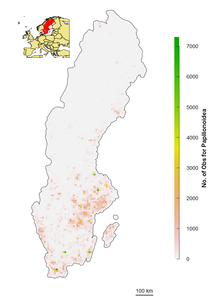
Amp=Amphibians; MamLnB=Land Mammals; Bir=Birds; Odo=Odonata; Opi=Opilionidae; Pae=Papilionoidea; Vas=Vascular Plants