|
Biodiversity Data Journal : Software description
|
|
Corresponding author: Carlo Allocca (carlo@hcmr.gr)
Academic editor: Viktor Senderov
Received: 18 Jun 2015 | Accepted: 13 Jul 2015 | Published: 29 Jul 2015
© 2015 Carlo Allocca, Alexandros Gougousis.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Allocca C, Gougousis A (2015) A Preliminary Investigation of Reversing RML: From an RDF dataset to its Column-Based data source. Biodiversity Data Journal 3: e5464. doi: 10.3897/BDJ.3.e5464
|

|
Abstract
Background
A large percentage of scientific data with tabular structure are published on the Web of Data as interlinked RDF datasets. When we come to the issue of long-term preservation of such RDF-based digital objects, it is important to provide full support for reusing them in the future. In particular, it should include means for both players who have no familiarity with RDF data model and, at the same time, who by working only with the native format of the data still provide sufficient information. To achieve this, we need mechanisms to bring the data back to their original format and structure.
New information
In this paper, we investigate how to perform the reverse process for column-based data sources. In particular, we devise an algorithm, RML2CSV, and exemplify its implementation in transforming an RDF dataset into its CSV tabular structure, through the use of the same RML mapping document that was used to generate the set of RDF triples. Through a set of content-based criteria, we attempt a comparative evaluation to measure the similarity between the rebuilt CSV and the original one. The results are promising and show that, under certain assumptions, RML2CSV reconstructs the same data with the same structure, offering more advanced digital preservation services.
Keywords
Reverse Mapping Language, RML, Linked Open Data, Data Preservation
Introduction
To date, a large percentage of scientific data published on the Web of Data (
Based on a set of content-based criteria to measure the similarity between the original data source and the one reconstructed by RML2CSV, we evaluate the approach over a collection of real-world RDF datasets from Biodiversity domain available in the MedObis repository (
The paper continues as in the following: The Study Area Description section briefly describes the underlying R2RML and RML mapping languages, demonstrating how they work in practice for exposing a CSV data source as an RDF dataset, and introduces the reverse process. It also details the main assumptions under which we analyse and develop the reverse process. The Design Description and Implementation sections describe the RML2CSV algorithm and its implementation. The Evaluation and Results section defines the main criteria to evaluate the approach and details the results. The Discussion section discusses upon the achievements and propose a number of solutions for relaxing the two assumptions that we will be part of future development. The Related Works section discusses relevant works. Finally, Conclusion and Outlook section concludes the work describing the main achievements and provide a road-map for future work.
Project description
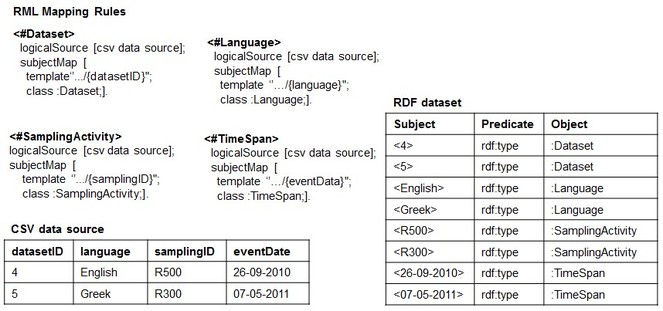
First, we briefly introduce the R2RML and RML ([R2]RML) mapping languages to the extent at which it concerns with our preliminary investigation (see (
An R2RML and RML Overview. R2RML provides a declarative language for expressing customized mappings from relational database to RDF dataset, expressed in a structure and target vocabulary of the Engineer's mapping choice (
To face with the high expressivity of RML's mapping language and to monitor the complexity of the reverse process, we have finalised, implementation included, the current work considering a subset of RML: RML Lite. The main restrictions that RML Lite imposes to a triples map are:
1. given a mapping rule tmi, the subject map is characterized by one template property and one class property with IRIs values,
2. given a mapping rule tmi, the predicate-object-Map references to one predicate property with IRI value and one object map which, in turn, is represented by an objectMap property with one value of referencing object map type,
3. given a mapping rule tmi, a referencing object map is represented by a resource that has exactly one parentTriplesMap property where the value must be a triples maps as defined above, known as the referencing object map's parent triples map,
4. if tm1,...,tmn are triples maps of the same RML Mapping Document and defined according to 1-3, they all refer to the same CSV data source.
Basically, RML Lite allows only the mapping of CSV columns to Class or Object Property of an RDF data model and, at the same time, it is expressive enough to discuss potential issues related to the reverse process in general, and how we intend to approach them. An example of RML Lite Mapping document is showed in
The CSV2RDF and RDF2CSV Processes. Generally speaking, mapping process aims at transforming instances of a data source structure into instances of target schema, preserving the semantic and allowing the implementation of an automatic algorithm to perform such a transformation (
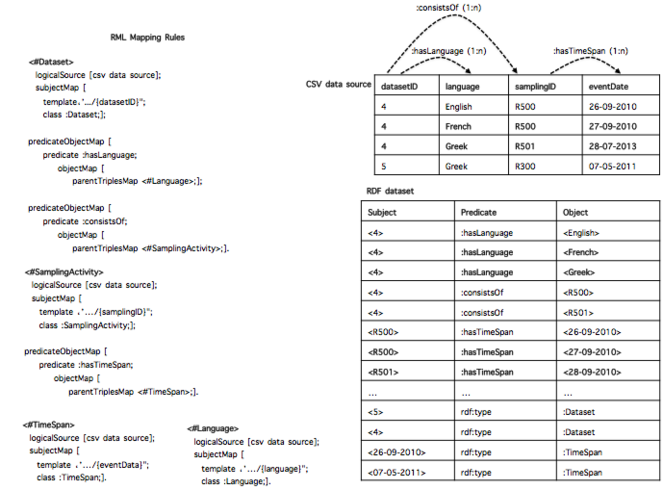
When using RML to perform such a task for a CSV data source (CSV2RDF), it means to write down a set of rules (stored as [R2]RML mapping document) that specifies how to semantically interpret both the structure and the data with respect to the target RDF data model. For example,
Conversely, RDF2CSV - the task of rebuilding the structure and the instances of the CSV data source from the RDF dataset - works in opposite direction: the RML rules are used to rebuild the column-based structure and populate it with the data from the RDF dataset. To exemplify, the rule <#Dataset>, when applied for the reverse process, retransforms the instances of the class: Dataset into values of the column datasetID.
Assumptions. The RDF2CSV process can pose a number of issues making it a very challenging task to accomplish. In what follow we present and discuss two of them: the first is related to the set of RML mapping rules used to expose the CSV data source in RDF and, the second concerns with the implicit cardinality constraints of the associations between the columns of the CSV data source. For both, in this preliminary study, we formulate assumptions to work with.
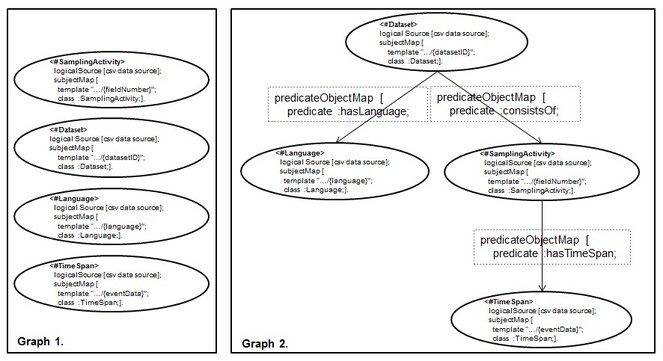
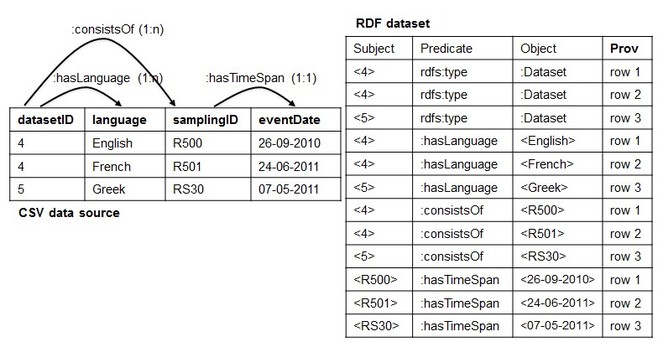
1. The Dependency Tree Assumption: It is related to the implicit structure that the set of RML mapping rules should form in order to succeed with the reverse process. Before formalizing it, we explain it by continuing the reverse of the RDF dataset of
What we have produced so far are only two dimensions (the columns and the cells) out of the three (the columns, the cells and the rows) that characterize a CSV data model.
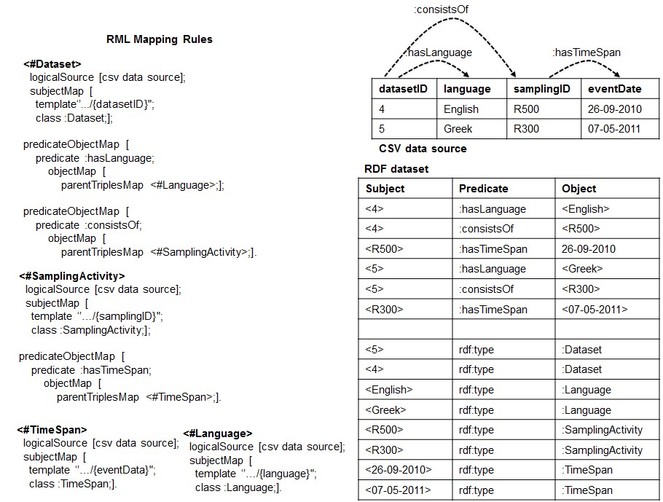
We noticed that the root of this problem may lie in the fact that potential relationships between columns in the CSV data source are not expressed at the conceptual level through the mapping rules. As shown in
Making explicit potential associations (guided by the target schema MarineTLO (
Based on such observation, we asked how we can make sure that we deal with types of scenario exemplified in
Dependency Tree Assumption (DTA). Given a set of RML mapping rules, S = {tm1,.. . , tmn}, that was used to expose a CSV data source, C, as an RDF dataset D. We use S over D to obtain back C if and only if the directed graph, G, underlying S is one n-ary tree.
Informally, G will have (a) only one vertice, root, that does not have incoming edges, (b) one or more vertices, leaves, that do not have outgoing edges, (c) there is at most one path (always starting from the root node) that connects two nodes and (d) each node has no more than n children.
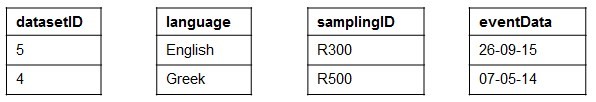
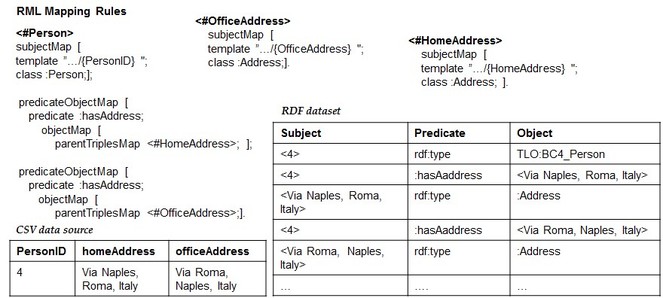
2. Implicit Cardinality Restrictions (ICR). It is related to the cardinality of the association between CSV columns. For the sake of clarification, let's consider the example of
Implicit Cardinality Restrictions (ICR). Given an RDF dataset D, we assume that D was generated from an original CSV data source C with associations between columns with only 0:0 or 1:1 cardinality constraints.
An example of CSV data source that satisfies the ICR assumption is showed in
Once the DTA and ICR are satisfied, the set of RML rules contains all the required information to rebuild the content, row by row, header included. In particular, each rule provides details such as the SubjectMap and PredicateObjectMap that connects two rules (e.g the predicate: consistsOf connects <#Dataset> with <#SamplingActivity>). Taking advantage of such structures, one way to build back a specific row is to exploit the set of rules from the most generic one to the most specific ones. Using a tree nomenclature, it means to visit the n-ary tree from the root to the leaves. We repeat this step for all the values that are instances of the root SubjectMap's Class. To exemplify the main idea, let us consider the RDF dataset and the set of rules of
Algorithm
To compute automatically such a process we devised a generic (as it provides the main steps that can be used also for other types of sources such as XML, JSON, DB) and extendible (as it provides the main logic for covering other RML language features as well as) algorithm, RML2CSV, as detailed in Algorithm 1. In particular, line 3 identifies the most generic triple map (it is the one that does not have any incoming edge) and line 4 retrieves the instances of the SubjectMap class of that triple map by using the SelectDistinctSubejct(classURI, d) function. The latter is based on the SPARQL query saves as PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> Select distinct ?subject Where { ?subject rdf:type classURI } executed over the RDF dataset. Finally, we use the set of RML rules to reconstruct all the rows (from line 5 to line 9) using the ReverseRow sub-call as reported in the Appendix. Once all the rows are reconstructed, line 10 exports and save them as csv file.
Algorithm 1. Reversing an RDF Dataset through the use of RML mapping rules.
INPUT: 1) a set of RML mapping rules S 2) an RDF Dataset d.
OUTPUT: 1) a CSV file.
1: procedure RML2CSV (S, d)
2: reversedCSV[] ← empty; //List of reversed rows.
3: dT ← IdentifyTheMostGenericRMLrule(S); //dT ← the root node.
4: distinctSubjects[] ← SelectDistinctSubject(dt.getClassURI(), d);
5: for each subji in distinctSubjects[] do
6: partRevRowi [] ← empty; //List of rowItem.
7: currPred ← empty; //a predicate of an RDF triple.
8: reversedRowi[] ← ReverseRow(S, subji , partRevRowi[], currPred, dT, d);
9: reversedCSV[].add(ReversedRowi[]);
10: Export reversedCSV[] as a csv text file.
The work has been supported by the LifeWatchGreece project, funded by GSRT, ESFRI Research Infrastructures, Structural Funds, OPCE II (Act Code: 384676).
Web location (URIs)
Technical specification
Repository
Usage rights
Implementation
Implements specification
We have implemented RML2CSV on top of RML, based on the fact that, in comparison to the other approaches, it provides a uniform way to access different types of data sources such as CSV, XML, JSON and DB. Consequently, we believe that enabling the corresponding reverse processes within the same framework it would not only strengthen the latter but also make it to be used by a much larger community, as well as to extend it to support other type of data source, beyond CSV. The current implementation of the RML2CSV can be found at https://bitbucket.org/carloallocca/rml2csv (see the three packages). It is important to highlight that the extension from RML Lite to RML does not have any logical implication on the presented algorithm. Moreover, we are currently working on it in order to cover the entire RML Mapping Language.
Additional information
Evaluation and Results
The general goal of evaluating RML2CSV is to answer the following (related) questions: 1. Does it solve the problem that is supposed to? 2. Does it work correctly under all the assumptions? To answer such questions, we designed a set of content based criteria to estimate the extent to which the reversed data source (csvr) overlaps, row by row, with the original one (csvo). To this end, we based such a comparison on computing a similarity measure between csvr and csvo, as expressed in the following:
\(ContentSimilarity(csv^{r}, csv^{o}) = 1 - contentDistance(csv^{r}, csv^{o}) \hspace{10mm}(1)\)
where the contentDistance intends to measure the number of rows and the extent to which they contain the same information. It is defined as in the following:
\(contentDistance(csv^{r}, csv^{o})= \sum\limits_{i=1}^m \frac{1}{m} \times rowDistance(row_{i}^{o}, row_{i}^{r}) \hspace{10mm}(2)\)
where m is the number of rows of the csvo, rowir is computed by CorrRow(rowio) which is a function to calculate the corresponding i-th row in the reversed CSV and, the rowDistance measures the number of cells and the extent to which they contain the same values. It is defined as in the following:
\(rowDistance(row_{i}^{o}, row_{i}^{r}) = \sum\limits_{i=1}^n \frac{1}{n} \times cellDistance(cell_{i}^{o}, cell_{i}^{r}) \hspace{17mm}(3)\)
where n is the length of rowio , cellsir is computed by CellRow(cellio) which is a function to calculate the corresponding i-th cell in the reversed CSV and the cellDistance is based on a string compare function checking whether the reversed value is the same as of the original one. Thus, cellDistance = 1, it means that the two values are different whereas cellDistance = 0 means that the two value are syntactically equal.
Combining (1), (2) and (3) together we have that: if (3) is always equal to 0, meaning that anytime we compare two rows they always contain the same values, then (2) is equal 0, meaning that csvr and csvo contain the same content. In this case, (1) would measure a similarity equal to 1. On the contrary, if (3) is always equal to 1, meaning that anytime we compare two rows they always contain different values, then (2) is equal 1, meaning that csvr and csvo contain different content. In this case, (1) would measure a similarity equal to 0. To face with the
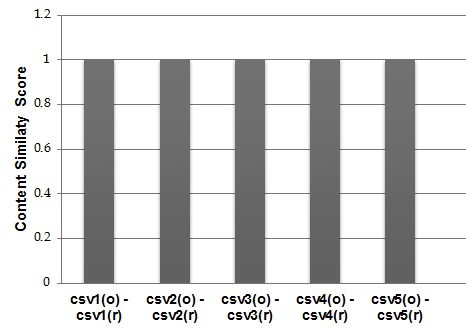
The current evaluation is based on a collection of five CSV data sources from Biodiversity domain, containing mainly occurrence data from the MedOBIS (Biogeographic information system for the eastern Mediterranean and Black Sea (Arvanitidis et al. 2006)). They are characterized by a different column-based structure containing from 4 to 12 columns (e.g. datasetID, language, fieldNumber, different types of measurements just to report a few). Before transforming them into RDF datasets we applied a pre-preprocessing to make sure that their content would not generate any of the issues analyzed in the Study Area Description section and further analyzed in the Discussion section. After running RML2CSV we compared csvr and csvo according to the criteria (1), (2) and (3). The results are shown in
As it can be noticed, RML2CSV reconstructed all the five CSVs with a content up to 100% overlapped with the original ones. This very initial evaluation does not pretend to demonstrate the correctness or completeness of proposed approach, but it posed the base and encourage us for a thorough evaluation of the RML2CSV efficiency and effectiveness.
Discussion
We designed and implemented our algorithm, RML2CSV, taking into account the DTA and the ICR assumptions. Now, we discuss how to build upon the current achievemnts in order to suggest solutions for relaxing the two assumptions.
More about the DTA and ICR: Being aware that they could be too limited for dealing with a wide range of real cases, we propose two solutions for relaxing the two assumptions. The first is based on extending the forward process producing an auxiliary structure for keeping links between RDF triples that refer to the subparts of the same row.
Mapping Quality Level:
Related Work
To the best of our knowledge, there is no other study investigating the reversing of an RDF dataset for reconstructing the original tabular data source of CSV type. On the contrary, several solutions exist to execute mappings from different types of data sources and serialisations to the RDF data model. The R2RML W3C recommendations (
In particular, RDF2RDB (
Similarly (
Unfortunately, all these existing approaches are rather limited for our scenario either because they do not consider the reverse problem at all or because they face it in different context and targetting diverse goal. While they contribute interesting elements for us to build on, we focus here on how to perform the reverse process for the case of column-based structured data source of CSV type w.r.t its original data structure and not any. Furthermore, as our solution is based on [R2]RML mapping language, it provides the additional advantage that we can perform both transactions, CSV data source to RDF dataset and vice-versa, within the same framework, that none of the discussed work does.
Conclusion and Outlook
In this paper we argue that an important aspect of long-term preservation of digital objects, such RDF datasets, is to provide full support for reusing such data, including mechanisms to bring back the data to their original format. To achieve this, in this work we investigated on how to perform the reverse process for the case of column-based data source such as tabular data. In particular, we devised an algorithm, the RML2CSV, for transforming an RDF dataset into its original data structure, through the use of the same RML mapping rules used to generate the set of RDF triples. The results of the evaluation showed that RML2CSV rebuilds the same data content with the same data structure under certain assumptions.
In the future, a thorough evaluation of RML2CSV efficiency will be performed. In addition, we have planned to extend RML2CSV to dealwith any type of constraints between columns (e.g. 1:n and, more general m:m) as discussed in Discussion section and to cover all RML mapping languages. As a long term objective, we plan to design and implement the back transformation to any type of relevant formats including XML, JSON and DB, by taking advantage of the achievements presented in this paper.
Appendix.
Algorithm 2 Reversing a single CSV row from an RDF Dataset through the use of RML mappings.
1: procedure ReverseRow(subji, partRevRowi [], currP red, dT, d)
2: currentRowContent[] ← partRevRowi [];
3: if dT = null then
4: currSubjectValue ← subji;
5: if dT.PredicateObjectMaps[] = null ∧ dT is not the root node then
6: return currentRowContent[];
7: if dT.PredicateObjectMaps[] = null ∧ dT is the root node then
8: termName ← dT.SubjectMap.getTemplate.localName;
. we need to check if SubjectMap has a template
9: rowItem ← TermName@currSubjectValue;
10: currentRowContent[].add(rowItem);
11: return currentRowContent[];
12: else
13: for i = 1 to dT .P redicateObjectM aps[].length do
14: currP red ← dT.PredicateObjectMap[i].getPredicate;
15: objectMap ← dT.PredicateObjectMap[i].getObjectMap;
16: if objectMap contains a parentTripleMap then
17: parentTriplesMap ← objectMap.getParentTriplesMap();
18: tripleMapName ← parentTriplesMap.getName();
19: nextTripleMap ← Search(dT, tripleMapName);
20: termName ← nextTripleMap.SubjectMap.getTemplate.localName;
. we need to check if SubjectMap has a template
21: className ← nextNodeToExplore.SubjectMap.getClass;
22: nextSubject ← SelectDistinctObject(d, currSubjectValue, currPred, className);
. SPARQL query where (currSubjectValue, currPred, ?object) and (?object rdf:type className)
23: cellItem ← termName@nextSubject;
24: currentRowContent[].add(cellItem);
25: ReverseRow(nextSubject, currentRowContent[], currPred, nextTripleMap, d);
26: if dT is the root then
27: termName ← dT.SubjectMap.getTemplate.localName;
28: rowItem ← termName@subji;
29: currentRowContent[].add(rowItem);
. if it is not already added
30: if objectMap contains a rr:reference then
31: print(”Not detailed for space reason.”);
32: return currentRowContent[];
33: else return null;
Acknowledgements
The authors would like to thank Nicolas Bailly for fruitful discussions and for providing valuable input to the issues of this article and Anastasia Dimou for contributing tremendously to the paper even though declining co-authorship. This research has been supported by LifeWatchGreece project (Funded by GSRT, EFRI Projects, Structural Funds, OPCE II).
References
-
A Direct Mapping of Relational Data to RDF. http://www.w3.org/TR/rdb-direct-mapping/. Accession date: 2015 5 15.
-
MedOBIS: Biogeographic information system for the eastern Mediterranean and Black Sea.,316.Marine Ecology Progress Series.225-230pp.
-
Linked Data - The Story So Far.International Journal on Semantic Web and Information Systems5(3):1‑22. DOI: 10.4018/jswis.2009081901
-
Supporting Tabular Data Characterization in a Large Scale Data Infrastructure by Lexical Matching Techniques.Communications in Computer and Information Science. URL: https://doi.org/10.1007/978-3-642-35834-0_5 DOI: 10.1007/978-3-642-35834-0_5
-
R2RML: RDB to RDF Mapping Language.W3C Recommendation 27 September 2012.25pp. URL: http://www.w3.org/TR/r2rml/
-
RDF Mapping Rules Refinements According to Data Consumers' Feedback.WWW Companion,Proceedings of the Companion Publication of the 23rd International Conference on World Wide Web Companion.249-250pp.
-
Extending R2RML to a source-independent mapping language for RDF.In Proceedings of the International Semantic Web Conference,Sydney, Australie.237-240pp.
-
Oracle XML DB Developer's Guide, 11g Release 2 (11.2).Oracle and/or its affiliates. All rights reserved.,54pp.
-
Some Preliminary Ideas Towards a Theory of Digital Preservation.DLF1-07,In Proceedings of the 1st International Workshop on Digital Libraries Foundations.34pp.
-
A generic reification strategy for n-ary relations in DL.OBML 2010 Workshop Proceedings24:67.
-
Linked Data: Evolving the Web into a Global Data Space.Synthesis Lectures on the Semantic Web: Theory and Technology1(1):1‑136. DOI: 10.2200/s00334ed1v01y201102wbe001
-
Jena - A Semantic Web Framework for Java.URL: http://jena.sourceforge.net/index.html
-
AquaMaps: Predicted range maps for aquatic species.ACM.12pp. URL: http://www.aquamaps.org/
-
Mapping Language for Information Integration.Technical Report FORTH-ICS/TR-385..15pp.
-
XLWrap – Querying and Integrating Arbitrary Spreadsheets with SPARQL.Lecture Notes in Computer Science. URL: https://doi.org/10.1007/978-3-642-04930-9_23 DOI: 10.1007/978-3-642-04930-9_23
-
Mapping Master: A Flexible Approach for Mapping Spreadsheets to OWL.Lecture Notes in Computer Science. URL: https://doi.org/10.1007/978-3-642-17749-1_13 DOI: 10.1007/978-3-642-17749-1_13
-
R2D: A bridge between the semantic web and relational visualization tools.IEEE,303-311pp.
-
R2D: Extracting Relational Structure from RDF Stores.2009 IEEE/WIC/ACM International Joint Conference on Web Intelligence and Intelligent Agent Technology,6pp. URL: https://doi.org/10.1109/wi-iat.2009.63 DOI: 10.1109/wi-iat.2009.63
-
Towards a Long-term preservation infrastructure for earth science data.Preservation of Digital Objects.89pp.
-
Scalable long-term preservation of relational data through SPARQL queries.Semantic Web Journal,SWJ '13.15pp.
-
Scalable Reconstruction of RDF-archived Relational Databases.Proceedings of the Fifth Workshop on Semantic Web Information Management - SWIM '13,1-4pp. URL: https://doi.org/10.1145/2484712.2484717 DOI: 10.1145/2484712.2484717
-
Model for Tabular Data and Metadata on the Web.W3C Working Draft 08 January 2015. URL: http://www.w3.org/TR/tabular-data-model/
-
Language-integrated querying of XML data in SQL server.Proceedings of the VLDB Endowment1(2):1396‑1399. DOI: 10.14778/1454159.1454182
-
A Transformation from RDF Documents and Schemas to Relational Databases.2007 IEEE Pacific Rim Conference on Communications, Computers and Signal Processing,15pp. URL: https://doi.org/10.1109/pacrim.2007.4313171 DOI: 10.1109/pacrim.2007.4313171
-
Conversion and emulation-aware dependency reasoning for curation services.Proc. 9th Int. Conference on Preservation of Digital Objects.iPres2012,38-45pp.
-
Integrating Heterogeneous and Distributed Information about Marine Species through a Top Level Ontology.Communications in Computer and Information Science. URL: https://doi.org/10.1007/978-3-319-03437-9_29 DOI: 10.1007/978-3-319-03437-9_29
Supplementary material
Download file (26.09 kb)