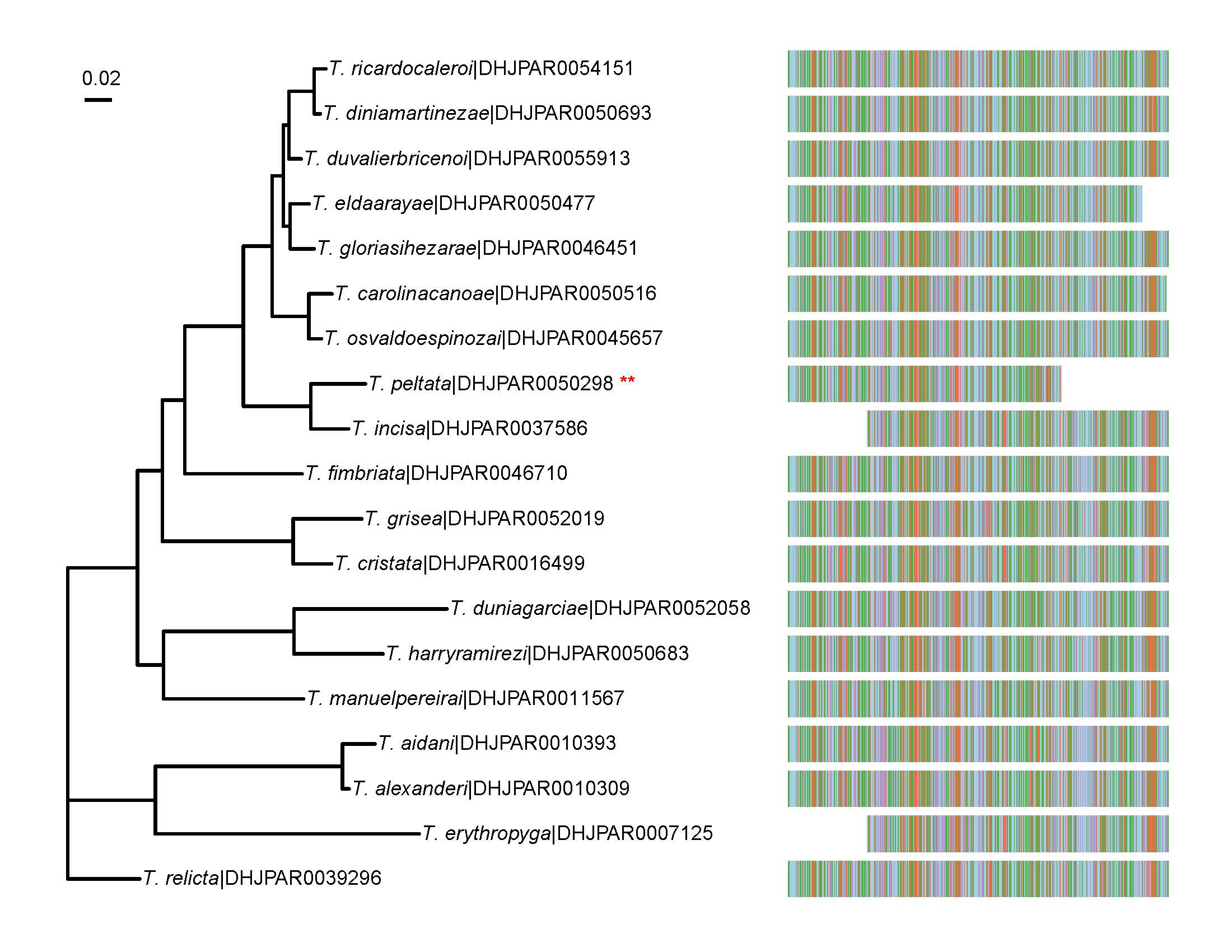
An unrooted phylogenetic tree for nineteen species of ACG Telothyria inferred by using the Maximum Likelihood (ML) method based on the General Time Reversible model (

|
||
|
An unrooted phylogenetic tree for nineteen species of ACG Telothyria inferred by using the Maximum Likelihood (ML) method based on the General Time Reversible model ( |
||
| Part of: Fleming A, Wood DM, Smith MA, Dapkey T, Hallwachs W, Janzen D (2020) Revision of Telothyria van der Wulp (Diptera: Tachinidae) and twenty-five new species from Area de Conservación Guanacaste in northwestern Costa Rica with a key to Mesoamerican species. Biodiversity Data Journal 8: e47157. https://doi.org/10.3897/BDJ.8.e47157 |