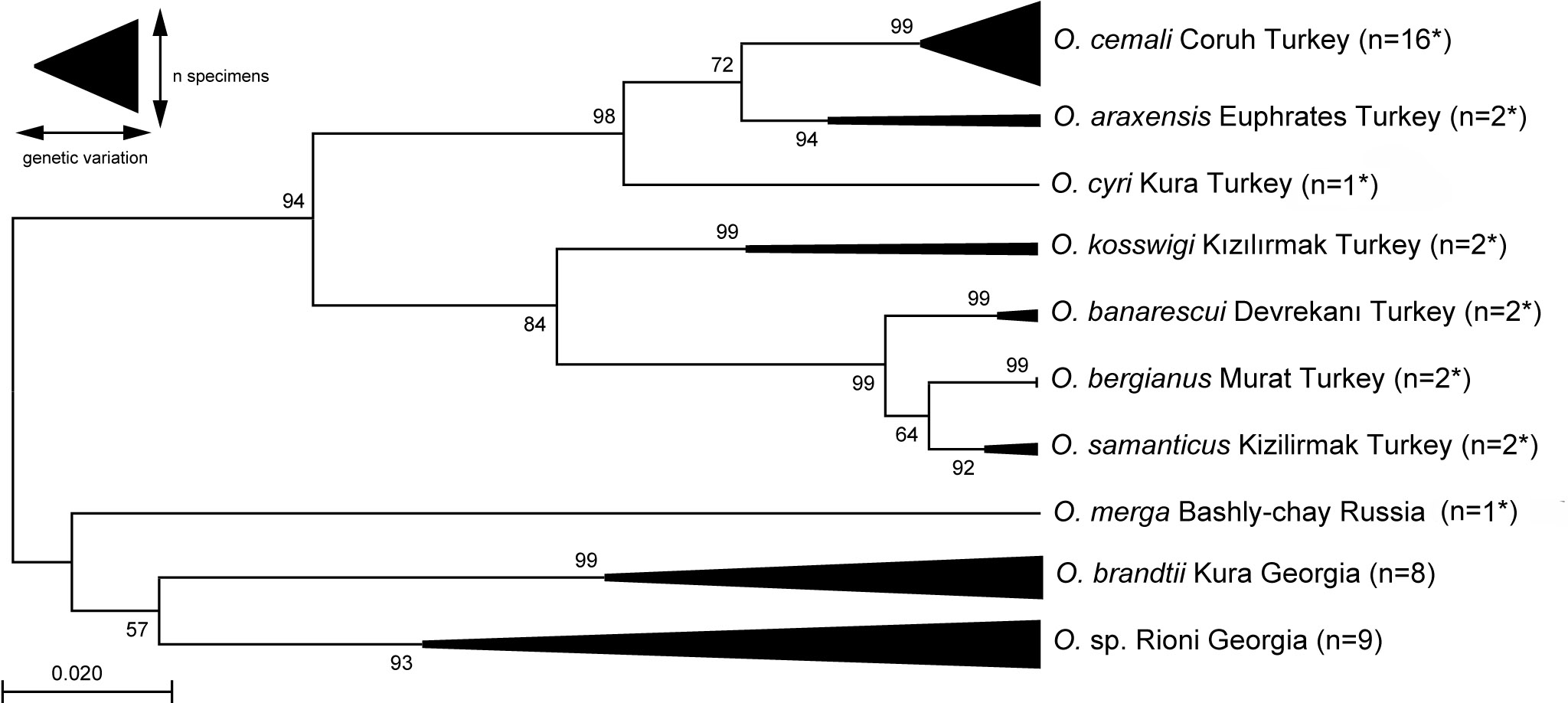
Maximum Likelihood estimation of the phylogenetic relationships of Oxynoemacheilus loaches, based on the mitochondrial COI barcode region (Kimura 2-parameter model, discrete Gamma distribution for rate differences with three categories + G parameter = 0.0610). Nucleotide positions with less than 95% site coverage were eliminated, resulting in 637 analysed positions. Numbers near nodes indicate bootstrap support values from 1000 pseudoreplicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis includes 45 nucleotide sequences taken from