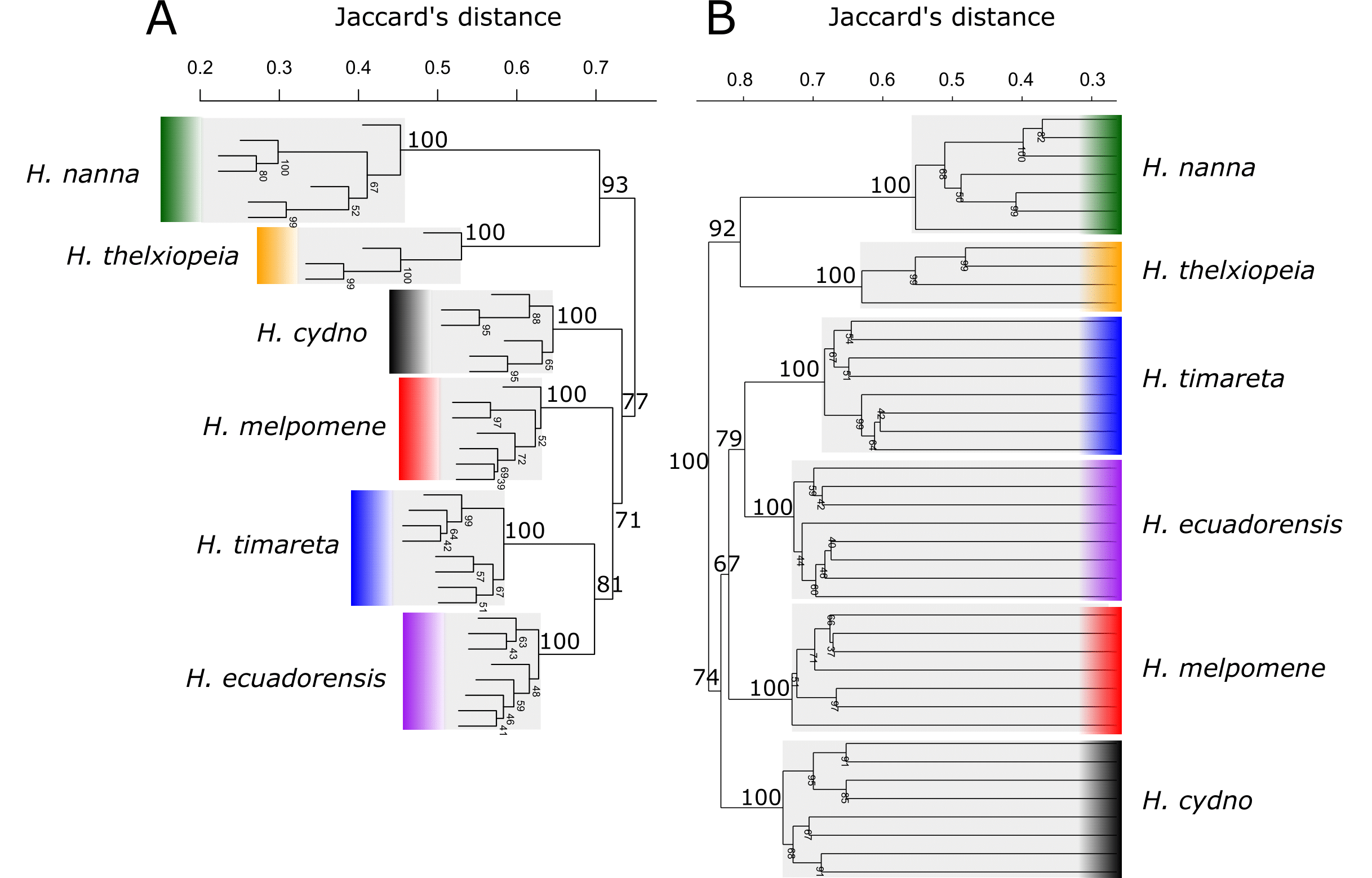
Comparison of hierarchical clustering trees in A) BinMat and B) PAST using the data taken from

|
||
|
Comparison of hierarchical clustering trees in A) BinMat and B) PAST using the data taken from |
||
| Part of: van Steenderen C (2022) BinMat: A molecular genetics tool for processing binary data obtained from fragment analysis in R. Biodiversity Data Journal 10: e77875. https://doi.org/10.3897/BDJ.10.e77875 |