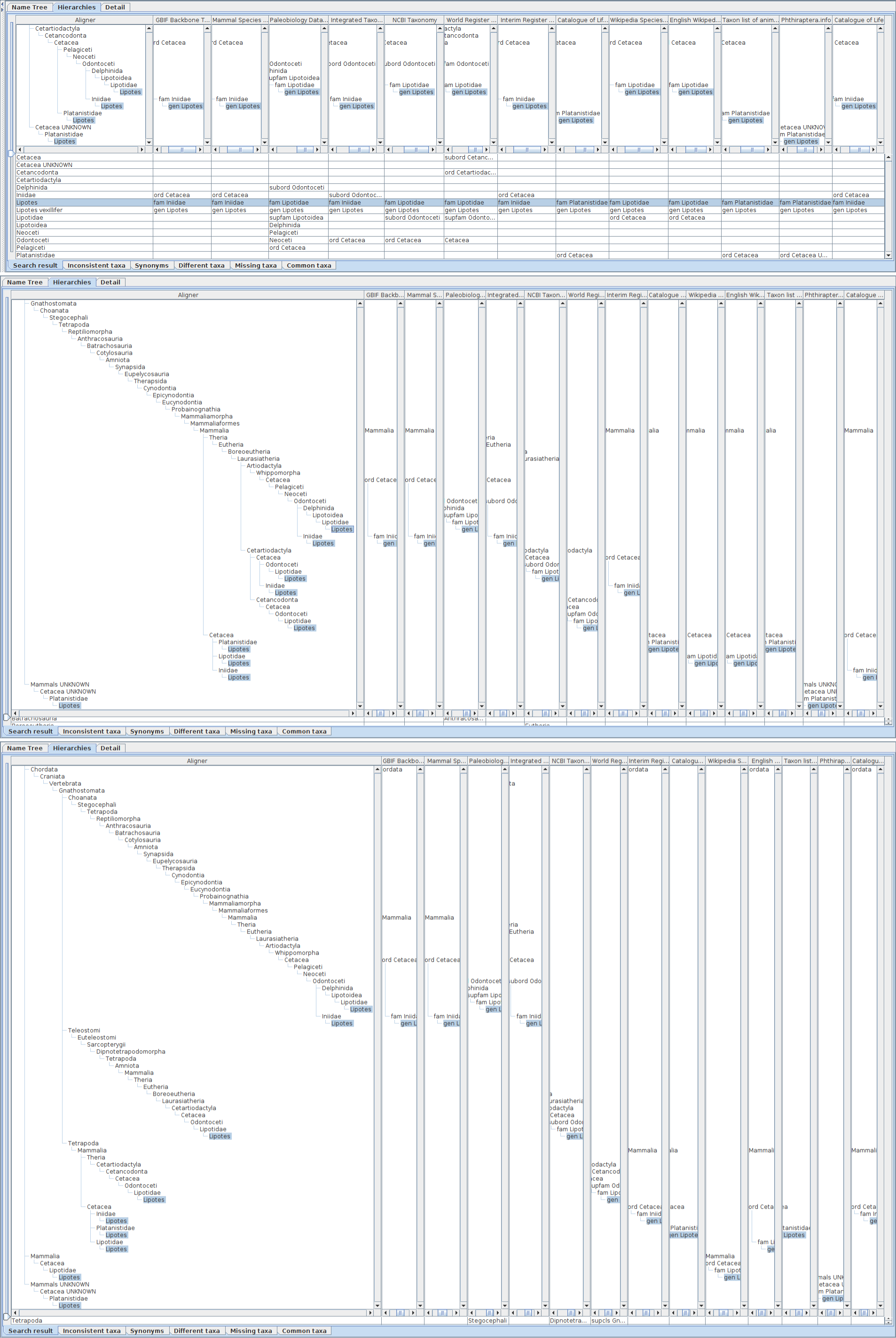
Alignments with different height of hierarchies containing Lipotes, up to order (upper), class (middle) or phylum (bottom), respectively. The height and depth of hierarchies being compared can be specified in the name usage list pane by either rank or number of levels above and below the name usage. If specified by rank, the software composes hierarchies to be limited within less than the next rank. For example, if class is specified as upper limit of hierarchies, the results may contain name usages less than phylum to cope with unranked intermediate name usages. The aligner tree for hierarchies up to order has four branches containing Lipotes. One branch is an artefact of misinterpreted data, Cetacea UNKNOWN, where UNKNOWN should be processed as authority rather than a part of name literal itself. Other three branches are results of different family names which require independent path between order and genus. The aligner tree for hierarchies up to class contains 9 Lipotes branches. Besides the erroneous UNKNOWN branch, remaining eight branches are joined into three higher branches depending on intermediate taxa between Mammalia and Cetacea, i.e. without intermediates, via Artiodactyla or Cetartiodactyla. Note that hierarchies without intermediates are mapped to branches in aligner tree either with or without intermediates depending on the sequence of integration into the aligner tree, because addition of an inconsistent path to the aligner tree requires the choice of one of those paths to which a new direct hierarchy is mapped but there is no way to choose one of them. The aligner tree up to phylum also contains 9 branches of Lipotes including one erroneous 'UNKNOWN'. Although the number of branches is the same to that for class, branching points get higher.