|
Biodiversity Data Journal :
Software description
|
Symbiota – A virtual platform for creating voucher-based biodiversity information communities
|
Corresponding author:
Academic editor: Lyubomir Penev
Received: 19 May 2014 | Accepted: 19 Jun 2014 | Published: 24 Jun 2014
© 2014 Corinna Gries, Edward Gilbert, Nico Franz
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Gries C, Gilbert E, Franz N (2014) Symbiota – A virtual platform for creating voucher-based biodiversity information communities. Biodiversity Data Journal 2: e1114. https://doi.org/10.3897/BDJ.2.e1114
|
|
Abstract
We review the Symbiota software platform for creating voucher-based biodiversity information portals and communities. Symbiota was originally conceived to promote small- to medium-sized, regionally and/or taxonomically themed collaborations of natural history collections. Over the past eight years the taxonomically diverse portals have grown into an important resource in North America and beyond for mobilizing, integrating, and using specimen- and observation-based occurrence records and derivative biodiversity information products. Designed to mirror the conceptual structure of traditional floras and faunas, Symbiota is exclusively web-based and employs a novel data model, information linking, and algorithms to provide highly dynamic customization. The themed portals enable meaningful access to biodiversity data for anyone from specialist to high school student. Symbiota emulates functionality of modern Content Management Systems, providing highly sophisticated yet intuitive user interfaces for data entry, batch processes, and editing. Each kind of content provision may be selectively accessed by authenticated information providers. Occupying a fairly specific niche in the biodiversity informatics arena, Symbiota provides extensive data exchange facilities and collaborates with other development projects to incorporate and not duplicate functionality as appropriate.
Keywords
Biodiversity informatics, digitization, economy of scale, natural history collection, Open Source, virtual collection portals
Introduction
For more than 250 years biodiversity has been documented in the form of specimens deposited, curated and maintained in over 1600 natural history collections in the United States. More than 1 billion U.S. specimens are widely used in scientific research, land management decision making and education (
Several approaches have been developed taking advantage of technologies for reducing repetitive work of digitizing collections and improving the quality of data. Each of the technologies currently in use and providing varying degrees of functionality for managing collections, digitizing specimens, providing on-line access to the records, or supporting large-scale analyses has been developed by and for different user communities and with slightly different emphases on the above mentioned functionalities; e.g., Arctos (http://arctosdb.org/), KE Emu (http://emu.kesoftware.com/), SilverBiology (http://www.silverbiology.com/), Specify (http://specifysoftware.org/), or VertNet (http://vertnet.org/).
This paper provides an overview of one such framework, the Symbiota software platform for networking biodiversity data (
In what follows, we concentrate on Symbiota's overarching principles, functions, and current and future applications. More detailed information on the functionality of particular modules (such as the interactive keys) will be treated elsewhere. We begin with reviewing the history of Symbiota's origins which has shaped the overall design with regards to conceptual, technical, and sociological aspects. We then discuss modules for managing specimen and/or observation occurrences, biotic inventories, and identification keys each from the perspective of general portal users and contributors. We close with an overview of Symbiota's current use and acceptance. Throughout, we attempt to explain why Symbiota has been embraced by its user communities, and lay out design and functionality trade-offs with relevance to larger-scale trends in biodiversity informatics (
Symbiota design principles – history and conceptual aspects
The history of pre-Symbiota development dates back to early digitization efforts and local solutions for database supported collections management. Early technical collaborations involved the HyperSQL project (
An impactful award made in 2008 led to an increasing separation from the initial general data management of Symbiota, initially as an independently designed MySQL/PHP identification tool working off of user-generated checklists. Subsequent redesign and enhancements led to the integration of a Darwin Core-based (
In its current realization, Symbiota is designed to support and promote grassroots bio-collaborations that work towards efficient data mobilization, improving data quality, and describing biodiversity in the form of virtual floras and faunas. Of particular importance is the emphasis on mobilizing information in order to address specific research questions, where the primary data are distributed among numerous collections. Collections are regarded as scientifically interdependent. The need to address shared research objectives takes primacy in software development over the more traditional design focus on customizable collection administration (e.g., handling in-house locations and curation trajectories of physical resources or processing loans). Although the focus on strengthening small grassroots collections collaborations still characterizes Symbiota's primary niche in the biodiversity data environment today, its modular design has enabled Symbiota to nimbly respond to changing needs and requirements, now supporting entire specimen digitization and collections management workflows plus extensive data exchanges with other systems.
Collaboration eases individual efforts and leads to enhanced information quality. Symbiota's primary focus is on permanently vouchered occurrence records of high scientific quality and accountability and on providing the tools to collaboratively manage this information. The platform has a modular framework for publishing biodiversity information; i.e., natural history collection occurrence records or observations, taxonomic information, images, species profiles, and taxon character and character state information in interactive keys for identification. Access to modules is achieved by emulating the functionality of a modern content management system (CMS). The system promotes a positive feedback loop that includes: (1) making data public instantaneously which can serve to expose errors; (2) using web messaging to alert responsible parties to such errors; (3) using web-based editing tools and workflows that allow such errors to be resolved as they are identified; (4) redirecting data repairs back to a source collection's internally used platform; and (5) rendering repairs permanent at the broader scale with the subsequent data update. In short, Symbiota leverages a themed, collaborative biodiversity data mobilization approach towards improving the quality of individual collections' data (
The CMS approach enables assignment of specific user permissions to share the tasks of data build-up and management of central resources. For instance, designated taxonomy coordinators may be responsible for the taxonomic thesaurus (see details below) to retain community-level acceptance and assure regular updates fostering the collaboration between taxonomic and regional experts. Symbiota also facilitates extensive data exchange options with other systems. For instance, collections record data may routinely be provided to the Global Biodiversity Information Facility (GBIF), taxon descriptions can be exchanged with the Encyclopedia of Life (EOL), and data may be synchronized with local management systems.
Portals encourage participation through shared branding and diversified forms of engagement. Symbiota can leverage and enrich biodiversity data through the creation of portals led by thematically coherent research communities. These communities tend to have shared taxonomic and/or geographic concentrations; and consequently shared interests to work collaboratively towards virtual floras and faunas that constitute authoritative, high quality vouchered treatments of a region's biota. In tailoring towards such communities, Symbiota has become a biodiversity information platform which is configurable, customizable, and independently manageable by each research community, and thus analogous to the Scratchpads approach (
Each web-based portal allows a wide range of forms of engagement for consortia, institutions, collections, research teams, individual researchers, and public groups or citizens (Fig.
Partial screenshot of the Search Collections panel for SEINet, showing Symbiota's ability to integrate the identities of individual member collections (with their respective logos and links to portal-configured homepages), regional portals (only four out of nine portals shown here), and multi-portal 'hubs'. All screenshots used in this paper were taken in February 2014.
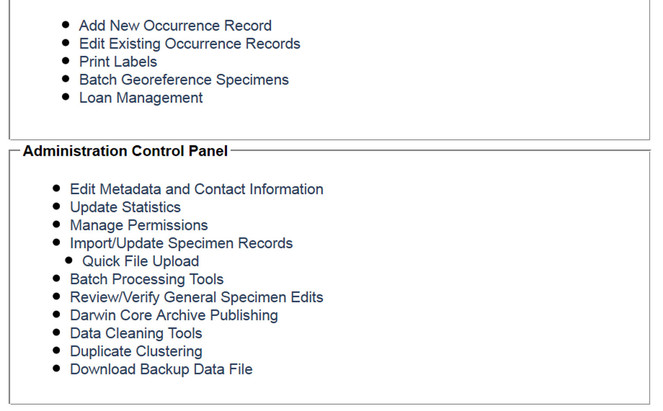
Screenshot of the Collection Management Panel for administrators of the University of Wisconsin Lichen Collection. The Data Editor Control Panel facilitates most day-to-day tasks related to specimen digitization as well as loan activity, whereas the Administration Control Panel focuses on managing the collection's appearance in the portal.
Symbiota furthermore allows individual collections to partake in multiple portals with overlapping thematic orientations. For instance, Arizona State University's Vascular Plant Herbarium is a member of both SEINet (Live Data) and Cooperative Taxonomic Resource for American Myrtaceae portal (CoTRAM; Data Snapshot), contributing only a subset of its full dataset to the latter. The flexibility offered in shaping and redirecting a collection's multiple virtual portal identities and thereby prioritizing one or more distinct biodiversity themes can enhance multi-portal growth and connectivity with minimal added effort.
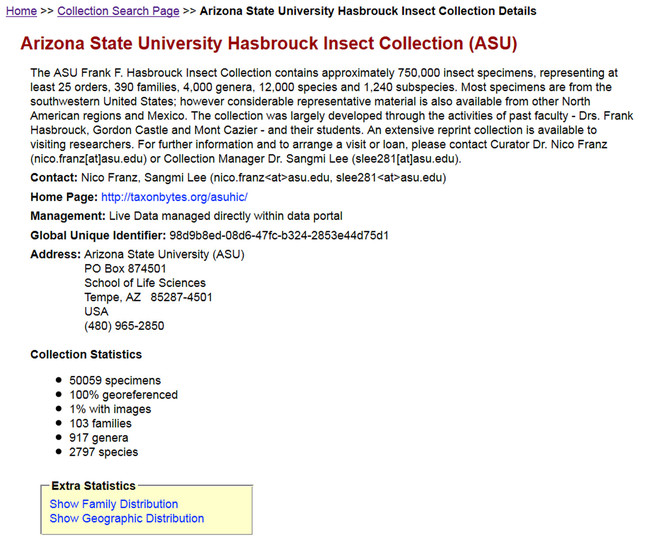
Although the collaborative approach is emphasized, portals maintain the provenance of information and content authorship. All occurrence records and images in Symbiota are indelibly tagged to their source collection, via a collection code and/or specific icon. Each collection, in turn, maintains a separate portal identity and homepage that provides a summary of its holdings, members, additional contact information, and collection statistics – number of specimens, percentage of georeferenced specimens, images, number of species, etc. (Fig.
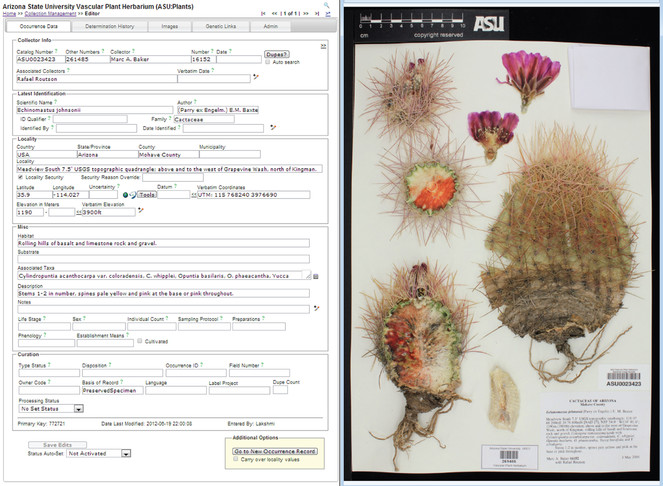
Modularization facilitates customization while maintaining data integrity. Symbiota strikes a balance between allowing portal communities to acquire distinct identities and functions while ensuring database integrity and consistency. The aim to network and integrate biodiversity data meaningfully sets limits to the degree to which portal configurations can vary. These limitations are most apparent in the format of the single occurrence data table (Fig.
Screenshot of the occurrence data form corresponding to the SEINet specimen ASU0023423 – Echinomastus johnsonii (Parry ex. Engelm.) E.M. Baxter (common name: Johnson's fishhook cactus – pertaining to the ASU Vascular Plant Herbarium collection. This is the primary Symbiota contributor interface through which individual specimen records are entered and edited. At the highest level, contributors can switch from the Occurrence Data tab to the Determination History, Images (see also right half), Genetic Links (e.g. to GenBank; Benson et al. 2012), and Administrative tabs. Entry personnel can zoom in on the label. Many data fields have inherent auto-completion or uniqueness/compliance checking functions, or can be expanded via the +-pencil icon for more fine-scale data entry. The Latest Identification data field section is linked to the taxonomic thesaurus, thereby ensuring correct integration of the Scientific Name with the portal-level taxonomy. Georeferencing support tools including Google Earth mapping and an embedded GEOLocate module. Under Curation, the Processing Status may be set to (e.g.) "Pending Review" to support filtering and data quality control practices among collection members.
Beyond sharing core biodiversity data formats and tables, Symbiota portals are customizable in numerous ways that suit specific performance and engagement needs of the corresponding communities. The concept of modularity applies to both the process of developing the software and the actual application instances. This approach promotes interoperability and extensibility. Modularity is manifested at different levels, as follows: (1) Application modularity – the modules for managing specimens, biotic inventories, identification keys, and species profile pages are designed to function independently of one another. Additional modules such as those supporting label image transcription (e.g., crowd sourced transcription, Optical Character Recognition, and Natural Language Parsing) or remote specimen identifications are turned on or off according to a portal's needs. Custom front-end frames, logos, texts, rotating images, and interactive identification games represent additional configuration options. (2) Data modularity – a portal's member collections are represented as independent units, each with its own management regime (Fig.
Modularization also means deploying existing web services and workflows that are optimized for specific functions instead of creating them anew. Symbiota subscribes to the 'small pieces loosely joined' tenet (
Technical design aspects
Symbiota adheres to the Open Source paradigm (http://opensource.org/osd.html; see also
Symbiota is exclusively web-based, which means that all portal data as well as management and user functions are accessible from any stationary or mobile device with a modern web browser and internet access. The software design philosophy parallels that of a Content Management System (e.g., Drupal or MediaWiki) specifically for the biodiversity community. If development of Symbiota began today a system like Drupal could be used as a starting point to implement Symbiota's functionality, however, this was not available when development on Symbiota started. Symbiota's central data model and algorithms implement principles of inheritance, hierarchy, and encapsulation, as found in object-oriented programming and ontology research (
Symbiota uses a centralized, server-based infrastructure solution for multiple parallel or inter-dependent information communities. The centralized model enhances performance and furthermore answers directly to the needs of many natural history collections; in particular medium- or small-sized collections with limited access to IT personnel and infrastructure (
Using Symbiota portals
In this section we describe functionality that any user without login credentials may access in a Symbiota portal (see also
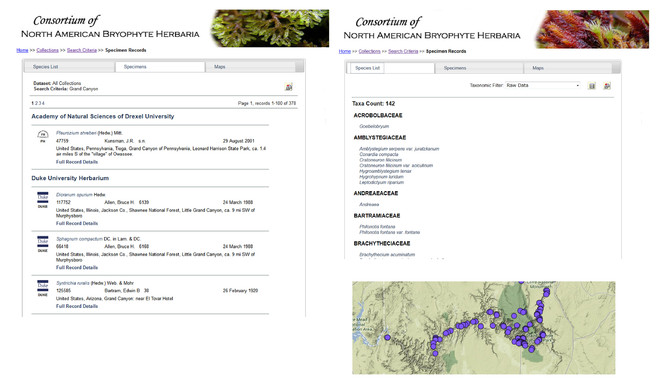
Dynamic flora and fauna checklists. Symbiota is designed to replace traditional, static floras or faunas with a dynamic approach. Modules are provided for each information concept found in such tomes – checklists, keys, taxonomic treatments and taxon descriptions with distribution maps and images. Linkages between the information content of each module provides for dynamic and user-driven data retrieval. Natural history specimen and (where appropriate) observation records are the core of any Symbiota portal, stored centrally in a relational schema based on the Darwin Core standard (http://www.tdwg.org/standards/450/). A standard specimen search engine with auto-completion and pick-list functionality facilitates taxonomic and geographic searches. Returned is a list of specimen records that match the criteria (Fig.
Screenshot of results displayed based on the search criteria ‘Grand Canyon’ which returns 378 records in 142 taxa. The Specimens panel (center tab) shown on the left presents an abbreviated summary view for each record that is expandable (not shown here). A Species List (left tab), shown on the right and Maps view (right tab, unselected), shown on bottom, are also available. A data icon in the top right corner facilitates downloading of the entire search results in Darwin Core or Symbiota CSV text file format (no permissions required). High-density occurrences of records are 'integrated' at coarser geographic scales and become resolved into separate latitude/longitude points at finer levels.
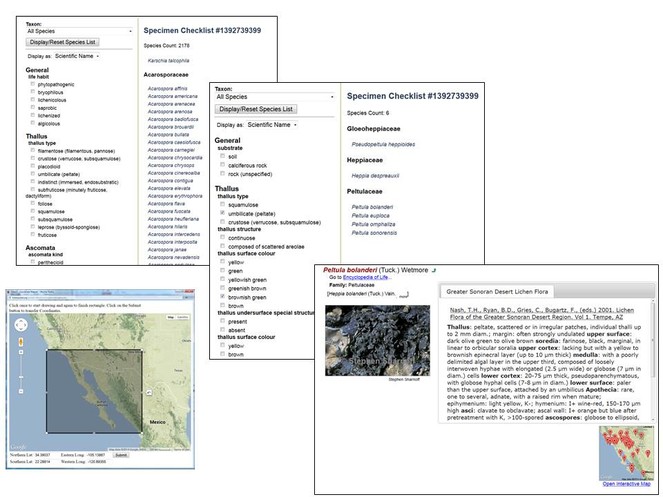
The main feature of the interactive keys is the capability to dynamically assemble character and character state information for any given taxon group combination. The module can handle vastly different sets of characters for disjointed taxonomic groups within a single dataset. It furthermore optimizes displaying only those characters and states that differentiate taxa under consideration at any given step in the identification process. With every decision a user makes during the keying process, characters and states are added and/or removed, starting with more generally applicable characters and gradually moving towards specialized traits (Fig.
Screenshots of several stages in an exemplary sequence of using the Dynamic Key function in Symbiota. (A) Using the Dynamic Map interface, a rectangle is selected; the coordinates correspond to the bounding box are selected. Users can restrict the subsequent checklist creation process ("Build Checklist") through specification of a Taxon Filter (i.e., higher taxa down to family, listed alphabetically). (B) Using these search criteria, the module will search vouchers in Symbiota that may satisfy these conditions, and integrate the pertinent voucher list into a taxon (family/species) list with 2178 species-level matches. The Dynamic Key interface (left menu) is initially simple, including choices regarding e.g. habit. (C) In this example, selecting only two criteria from the list will (1) reduce the count of taxon matches to 6 and (2) display a remaining taxa-specific list of traits suitable for further determination, drawn from the character inheritance hierarchy of the corresponding key in Symbiota. These can either be further scrutinized with the dynamic key or determined through viewing of individual species profile pages.
Symbiota provides attractive taxon profile pages with natural language descriptions as well as images for each taxon. An occurrence map generator utilizes information from the collections records to dynamically produce Google distribution maps. Links to external resources (such as the Encyclopedia of Life) and Google searches for additional web resources are supported.
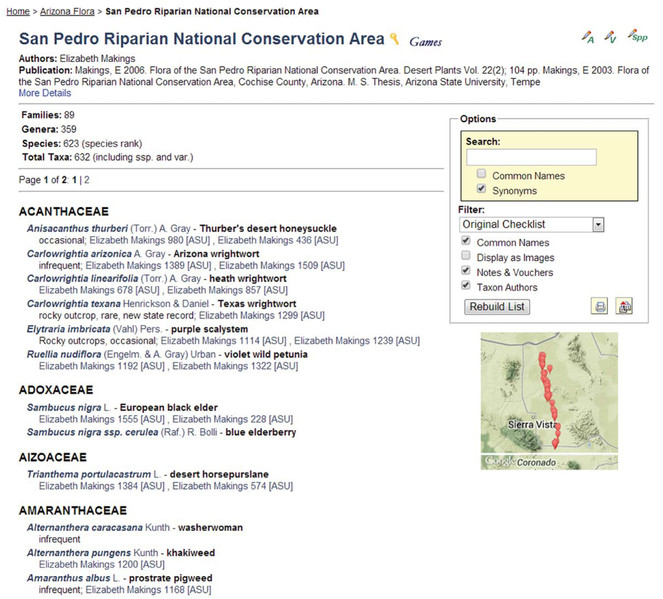
The concept of virtual floras and faunas is best realized through Symbiota's biotic inventories. Such inventories are usually developed by an expert or an expert team, frequently starting with or leading up to a published taxon checklist. While these lists (and, hence, floras and faunas) may be generated dynamically from occurrence records (Fig.
Screenshot of the homepage of the SEINet-derived San Pedro Riparian National Conservation Area checklist, a 'member' checklist of the Arizona Flora biotic inventory 'family' (Makings 2006, 2014). Checklist administrator functions are available in the top right corner ("A" – Administration, "V" – Manage Linked Vouchers, "Spp" – Edit Species List). The selected screenshot shows the entire list in alphabetical order, Taxon Authors, Common Names, and Notes & Vouchers. Clicking on vouchers listed for each taxon will display specimen details including images, when available. A Google Map thumbnail can be clicked for a more expansive map view.
In contrast to traditional print publications, these on-line checklists are fully rooted in voucher data records. A suite of management tools are available to build checklists, link vouchers, and coordinate species lists with herbarium level specimen annotations that result in the correction of misidentified vouchers. Maintenance of a set of checklist is facilitated by the ability to organize lists in a hierarchical relationship where parent checklists automatically inherit taxa and vouchers from all children checklists. This is particularly useful for creating integrated state and county lists; e.g., as currently in use by Park Service employees in the southwestern United States. Mangers can keep track of area-specific vouchers without the need of explicit collection level permissions from each herbarium holding the specimens. The hierarchical relationship of the checklists results in consolidated lists and reports available to the regional managers that are based on the independent efforts of a network of local data managers.
Contributing to and maintaining Symbiota portals
Below we offer an overview of Symbiota features available to data contributors who have been granted data editing rights beyond those of general users. The platform supports wide-ranging interconnections among management tasks, resources, modules, and the resulting floras and faunas produced by Symbiota portal contributors. Additional information regarding Symbiota's data management functions is available at http://symbiota.org/docs/symbiota-data-management-tools/. Each Symbiota module provides consistent editing interfaces employing intuitive icons and tabs.
Portal content management. User permissions allow for task-specific access control. For each portal, highest-level access rights are typically confined to a small group of portal-level super administrators who can assign initial rights to collection and project leaders. From that highest level, additional rights assignments can cascade down to the individual collection and project domains. This promotes arrangements where no single person has overwhelming responsibilities to manage rights, and no single-point bottlenecks are created in developing a portal's contributor community. 'Global' or 'local' portal leaders can manage rights of their collection and project members at the appropriate levels.
In the taxonomy managementmodule, portal contributors with appropriate taxonomy editor rights can edit names, add names, undertake taxonomic rearrangements, change existing or provide new synonymy information, eliminate names that are in error, and modify the locality security settings for sensitive taxa. Interactive and flexible batch upload functions are available which permit rapid population of the taxonomic thesaurus with large classifications rendered in compliant file formats. Typically, such mass taxonomy population tasks are assigned to the portal manager. The module also allows the development and selection of alternative mappings among valid names and synonyms, reflecting (e.g.) parallel classification schemes such as the Flora of North America (http://www.efloras.org/flora_page.aspx?flora_id=1) and the USDA Plants Database (http://plants.usda.gov/).
Symbiota's collection management module is appropriate for managing collections entirely on-line and inside the Symbiota portal framework. The module has two control panels to enable data editing and administration (Fig.
The data editor module supports workflows for digitizing label information from a specimen image. The label image (or label part of the image) is made available to the editor where Optical Character Recognition (OCR) and automatic parsing of this information into database fields (Natural Language Processing, NPL) may be applied. Voice recognition is currently in the experimental stage and may become available in the future for entering data. If a duplicate record was previously entered by a different institution, the system has the ability to quickly retrieve and import this information into the data entry form, thereby avoiding the need of retyping the same data for each duplicate specimen. This is a powerful feature for saving time and reducing errors in new entries as well as the existing one as they are collaboratively edited.
The administration control panel (Fig.
Biodiversity data products. Symbiota's interactive, multi-entry identification keys are generated directly from descriptive data stored in a relational data representation of the DELTA data standard (http://delta-intkey.com/). Such keys have many advantages over traditional, dichotomous keys (
Symbiota's novel integration of taxonomic and distributional information (Fig.
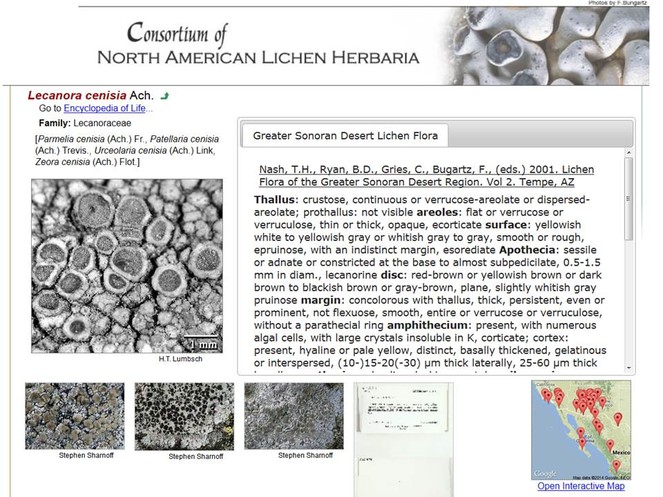
Taxon profile pages are the central vehicle in Symbiota for conveying information on a given taxon (Fig.
Screenshot of the Species Profile page of Lecanora cenisia. Ach. (Lecanoraceae) in CNALH (http://lichenportal.org/portal/taxa/index.php?taxauthid=1&taxon=53780&cl=2), with an outlink to the corresponding Encyclopedia of Life page (http://eol.org/pages/196725), common names, an assortment of accredited and clickable thumbnail images including in situ photographs and herbarium voucher scans (additional images and links are displayed when clicking links below), an Interactive Map for documented species occurrences, and taxonomic diagnosis (based on Nash et al. 2001 – also linked).
Finally, biotic inventories are products generated by experts or expert teams and may integrate information entered, edited, and maintained in all other modules towards a regional flora, fauna, or both. Dynamically generated as well as static species checklists may be included. Static species lists give researchers complete and continuous control over species composition, taxonomic placements, region specific comments, and voucher assignments. One of the benefits of a virtual flora or fauna environment is the ability to establish all-inclusive species lists as well as a multitude of smaller lists covering more limited regions within the overall area of study.
Current use and acceptance
As of May, 2014, 18 Symbiota portals are live and actively maintained on-line, representing more than 350 individual collections and a wide range of organismal groups (Table
List of Symbiota portal names and themes (in alphabetical order), URLs, and numbers of participating portal collections. Numbers generated in mid May, 2014.
|
# |
Portal Name and Theme |
Portal URL |
Collections |
|
1 |
CNABH – Consortium of North American Bryophyte Herbaria |
55 |
|
|
2 |
CNALH - Consortium of North American Lichen Herbaria |
58 |
|
|
3 |
CNH – Consortium of Northeastern Herbaria Portal |
31 |
|
|
4 |
CoTRAM – Cooperative Taxonomic Resource for Amer. Myrtaceae |
6 |
|
|
5 |
Herbario Virtual Austral Americano |
8 |
|
|
6 |
IRHN – Intermountain Region Herbarium Network* |
11 |
|
|
7 |
MABA – Madrean Archipelago Biodiversity Assessment – Fauna |
12 |
|
|
8 |
MABA – Madrean Archipelago Biodiversity Assessment – Flora* |
22 |
|
|
9 |
Macroalgal Herbarium Consortium Portal |
21 |
|
|
10 |
MyCoPortal - Mycology Collections Data Portal |
39 |
|
|
11 |
NANSH – North American Network of Small Herbaria* |
11 |
|
|
12 |
Neotropical Arthropod Portal |
5 |
|
|
13 |
Neotropical Flora Portal |
11 |
|
|
14 |
Northern Great Plains Regional Herbarium Network* |
21 |
|
|
15 |
SCAN – Southwest Collections of Arthropods Network |
21 |
|
|
16 |
SEINet – Southwest Environmental Information Network |
25 |
|
|
17 |
SERNEC - Southeast Regional Network of Expertise and Collections* |
7 |
|
|
18 |
Smithsonian Tropical Research Institute – Vertebrate Portal |
12 |
Many Symbiota portals are experiencing rapid growth in collection and occurrence record numbers, due in part to digitization projects supported by the NSF-ADBC program. Jointly these numbers and trends speak clearly of an increasing acceptance of Symbiota's bottom-up approach and flexibility in creating regionally or taxonomically themed biodiversity data communities.
Future directions
Symbiota plays a pivotal role in North America in mobilizing small- to medium-sized natural history collections to enable voucher-based biodiversity research. Beyond recent funding successes, its impact is most significantly manifested in the creation, maintenance, and continuous expansion of strong portal communities over many years. This success has been due both to Symbiota's integrated and stable web-based CMS and the ability to effectively promote and support self-motivating collaborations at various regional and taxonomic scales. The balance struck in establishing fixed practices for data integration while remaining responsive to specific needs of research teams is also important in this context. Symbiota maintains its niche in the biodiversity informatics realm through extensive collaborations and integration of functionalities developed by other projects, thus allowing its core modules to interact functionally and share data with many other systems.
Sustaining and growing this role is a challenge with scientific, technical, and socio-economic dimensions that will remain relevant as long as the primary sources of support are competitive innovation and research grants. Like any developing biodiversity data platform, Symbiota is bound by the requirement to invest into continuous, adequate information technology and personnel infrastructure to remain useful (
Because interconnecting taxonomic information is an essential part of Symbiota's advanced functionality, another scalability challenge is the management of taxonomic thesauri over time and across regions. Alternative taxonomic perspectives may receive endorsement from separate portal communities whose areas of coverage overlap at least in part. Such situations are well illustrated by a succession of floristic treatments in the Southern and Mid-Atlantic United States (
Readers interested in establishing new portals or joining existing portals can find more detailed, step-by-step instructions on the Symbiota software project website at http://symbiota.org.
Web location (URIs)
Technical specification
Usage rights
GNU General Public License, Version 2
Acknowledgements
The authors wish to thank (in alphabetical order) Mary Barkworth, Benjamin Brandt, Steven Buckley, Leslie Landrum, Peter McCartney, Shelley McMahon, Paul Morris, Thomas Nash, Robin Schroeder, Thomas Van Devender, and numerous other contributors for their critical vision and contributions during the decade-long development of the Symbiota software platform. Countless contributions of curators, collection personnel, students, and the greater user community are also kindly acknowledged. The development of Symbiota and advancement of related specimen digitization activities have been supported by the National Science Foundation under awards DBI-9983132, DBI-0096982, DBI-0237418, DBI-0743827, DBI-0847966, DEB-1155984, EF-1115116, and EF-1207107. Additional contributions through regional sources have further supported the development of Symbiota components.
References
- New Push to Bring US Biological Collections to the World's Online Community.BioScience61(9):657‑662. https://doi.org/10.1525/bio.2011.61.9.4
- The SALIX Method: A semi-automated workflow for herbarium specimen digitization.Taxon62(3):581‑590. https://doi.org/10.12705/623.16
- Beagrie N, Eakin-Richards L, Vision T (2010) Business models and cost estimation: Dryad Repository case study. In: Rauber A, Kaiser M, Guenther R, Constantopoulos P (Eds) iPRRES2010 Vienna Conference Paper, September.Österreichische Computer Gesellschaft,Vienna,365-371pp. URL: http://wiki.datadryad.org/wg/dryad/images/4/47/IPRES2010_Paper37.pdf [ISBN978-3-85403-262-5].
- ActKey: A Web-Based Interactive Identification Key Program.Taxon54(4):1041. https://doi.org/10.2307/25065490
- eFloras: New Directions for Online Floras Exemplified by the Flora of China Project.Taxon55(1):188. https://doi.org/10.2307/25065540
- Why Are We Still Producing Paper Floras? 1.Annals of the Missouri Botanical Garden98(3):297‑300. https://doi.org/10.3417/2010035
- Provenance in Databases: Why, How, and Where.Foundations and Trends in Databases1(4):379‑474. https://doi.org/10.1561/1900000006
- Euler/X: a toolkit for logic-based taxonomy integration.arXiv1402:1992. URL: http://arxiv.org/abs/1402.1992
- VertNet: A New Model for Biodiversity Data Sharing.PLoS Biology8(2):e1000309. https://doi.org/10.1371/journal.pbio.1000309
- Biodiversity data should be published, cited, and peer reviewed.Trends in Ecology & Evolution28(8):454‑461. https://doi.org/10.1016/j.tree.2013.05.002
- A flexible computer program for generating identification keys.Systematic Zoology23:50‑57. https://doi.org/10.1093/sysbio/23.1.50
- A general system for coding taxonomic descriptions.Taxon29:41‑46.
- A comparison of matrix-based taxonomic identification systems with rule-based systems.IFAC Workshop on Expert Systems in Agriculture, International Academic Publishers,Beijing.215-218pp. URL: http://delta-intkey.com/www/expertid.htm
-
A comparison of interactive identification programs.URL: http://delta-intkey.com/www/comparison.htm
-
3I Interactive Keys and Taxonomic Databases.URL: http://imperialis.inhs.illinois.edu/dmitriev/index.asp
- The potential for computer-aided identification in biodiversity research.Trends in Ecology & Evolution10(4):153‑158. https://doi.org/10.1016/s0169-5347(00)89026-6
- Description of two new species and phylogenetic reassessment of Perelleschus O’Brien & Wibmer, 1986 (Coleoptera: Curculionidae), with a complete taxonomic concept history of Perelleschus sec. Franz & Cardona-Duque, 2013.Systematics and Biodiversity11(2):209‑236. https://doi.org/10.1080/14772000.2013.806371
- Franz N, Peet R, Weakley A (2008) On The Use Of Taxonomic Concepts In Support Of Biodiversity Research And Taxonomy. Systematics Association Special Volumes. https://doi.org/10.1201/9781420008562.ch5
- Perspectives: Towards a language for mapping relationships among taxonomic concepts.Systematics and Biodiversity7(1):5‑20. https://doi.org/10.1017/s147720000800282x
- Names are not good enough: reasoning over taxonomic change in the Andropogon complex.Semantic Web Journal – Interoperability, Usability, Applicability - Special Issue on Semantics for Biodiversity1:1. URL: http://www.semantic-web-journal.net/content/names-are-not-good-enough-reasoning-over-taxonomic-change-andropogon-complex
- Assembling a virtual Weevils of North America checklist with Symbiota – preliminary insights.12th Biennial Conference of Science and Management on the Colorado Plateau.5pp. URL: http://nau.edu/Merriam-Powell/Biennial-Conference/Proceedings/
- Symbiota – promoting bio-collaboration.Project website1:1. URL: http://symbiota.org/
- New developments in museum-based informatics and applications in biodiversity analysis.Trends in Ecology & Evolution19(9):497‑503. https://doi.org/10.1016/j.tree.2004.07.006
- Approaches to the study of urban ecosystems: The case of Central Arizona—Phoenix.Urban Ecosystems7(3):199‑213. https://doi.org/10.1023/b:ueco.0000044036.59953.a1
-
DiversityDescriptions (DeltaAccess): documentation of the information model.URL: www.diversityworkbench.net/OldModels/Descriptions/Model/2005-03-30/DModelDD19.html
- Overview of interactive keys.Online Publication,1pp. URL: http://kikforum.wordpress.com/2007/01/12/overview-of-interactive-keys-provided-by-gregor-hagedorn/
- Introduction to SDD (Structured Descriptive Data), TDWG 2004.TDWG,1pp. URL: http://www.tdwg.org/2004meet/paperabstracts/TDWG_2004_Papers_Hagedorn_4.htm
- Structured Descriptive Data (SDD) version 1.0, TDWG.TDWG,1pp. URL: http://www.tdwg.org/2005meet/TDWG2005_Abstract_26.htm
- Biodiversity Online: toward a Network Integrated Biocollections Alliance.BioScience63(10):789‑790. https://doi.org/10.1525/bio.2013.63.10.4
- A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1):16. https://doi.org/10.1186/1472-6785-13-16
- Thematic Collections Networks.Online PublicationURL: https://www.idigbio.org/content/thematic-collections-networks
-
SALIX, the Semi-automatic Label Information Extraction system.URL: http://daryllafferty.com/salix/
- SALIX, the Semi-automatic Label Information Extraction system.Canotia,6pp. URL: http://nhc.asu.edu/vpherbarium/canotia/SALIX3.pdf
- Making good use of SEINet.online publication,8pp. URL: http://swbiodiversity.org/portal/misc/MakingGoodUseOfSEINet.pdf
- Natural history collections as sources of long-term datasets.Trends in Ecology & Evolution26(4):153‑154. https://doi.org/10.1016/j.tree.2010.12.009
- Biology: The big challenges of big data.Nature498(7453):255‑260. https://doi.org/10.1038/498255a
- A Computational- and Storage-Cloud for Integration of Biodiversity Collections.In Proceedings of the 2013 IEEE 9th International Conference on e-Science.78 - 87pp. https://doi.org/10.1109/escience.2013.48
- SEINet: metadata-mediated access to distributed ecological data. LTER.DataBitsSpring:1. URL: http://databits.lternet.edu/spring-2003/seinet-metadata-mediated-access-distributed-ecological-data
- CAP LTER Fifth Annual Poster Symposium, Center for Environmental Studies.Tempe, Arizona,February 19, 2003. URL: http://sustainability.asu.edu/docs/symposia/symp2003/McCartney_et_al.pdf
- The Open Knowledge Foundation: Open Data Means Better Science.PLoS Biology9(12):e1001195. https://doi.org/10.1371/journal.pbio.1001195
- Semantic Annotation of Mutable Data.PLoS ONE8(11):e76093. https://doi.org/10.1371/journal.pone.0076093
- An alternative species taxonomy of the birds of Mexico.Biota Neotropica4(2):1‑32. https://doi.org/10.1590/s1676-06032004000200013
- HyperSQL: web-based query interfaces for biological databases.Thirtieth Hawaii International Conference on System Sciences.329-339pp. https://doi.org/10.1109/hicss.1997.663405
- Network Integrated Biocollections Alliance. A strategic plan for establishing a Network Integrated Biocollections Alliance.NIBA,24pp. URL: http://digbiocol.files.wordpress.com/2010/08/niba_brochure.pdf
- Frequently asked questions related to Advancing Digitization of Biological Collections (ADBC) solicitation.NSF solicitation11-055:1. URL: http://www.nsf.gov/pubs/2011/nsf11005/nsf11005.pdf
- NSF awards grants to Advance Digitization of Biological Collections.Press Release11:136. URL: http://www.nsf.gov/news/news_summ.jsp?cntn_id=121015
- NSF-Supported Research Infrastructure: Enabling Discovery, Innovation and Learning.NSF,148pp. URL: http://www.nsf.gov/news/nsf09013/nsf_09013.pdf
- Innovation and Information Infrastructure: Making Sound Investments for E-Research.Research Bulletin, Boulder, CO22:12. URL: https://net.educause.edu/ir/library/pdf/ERB1022.pdf
- ZooKeys, unlocking earth's incredible biodiversity and building a sustainable bridge into the public domain: from "print-based" to "web-based" taxonomy, systematics, and natural history. ZooKeys Editorial Opening Paper.ZooKeys1:1‑7. https://doi.org/10.3897/zookeys.1.11
- Alternate Species Concepts as Bases for Determining Priority Conservation Areas.Conservation Biology13(2):427‑431. https://doi.org/10.1046/j.1523-1739.1999.013002427.x
- Darwin Core Archives – How-to Guide, Version 1, Released on 1 March 2011.Global Biodiversity Information Facility.,Copenhagen,23pp. URL: http://www.gbif.org/resources/2552
- GEOLocate.3.22.Belle Chasse, LA, Tulane University Museum of Natural History.. URL: http://www.museum.tulane.edu/geolocate/default.html
- Schoeninger R, Gries C, Nash TH (2002) Herbarium databases: creation and maintenance. Bibliotheca Lichenologica.82.291-299pp. URL: http://www.schweizerbart.de/publications/detail/isbn/9783443580612/Bibliotheca_Lichenologica_Band_82_Llimona_Lumbsch_ [ISBN3443580610].
- Rationale and value natural history collections digitization.Biodiversity Informatics7:77‑80. URL: https://journals.ku.edu/index.php/jbi/article/view/3994
- Scratchpads 2.0: a Virtual Research Environment supporting scholarly collaboration, communication and data publication in biodiversity science.ZooKeys150:53. https://doi.org/10.3897/zookeys.150.2193
- Stahlecker T, Kroll H (2013) Policies to Build Research Infrastructures in Europe – Following Traditions or Building New Momentum? Working Papers Firms and Region No. R4/2013.Fraunhofer ISI – Institute for Systems and Innovation Research.URL: http://www.isi.fraunhofer.de/isi-media/docs/p/de/arbpap_unternehmen_region/ap_r4_2013.pdf
- The Value of Museum Collections for Research and Society.BioScience54(1):66. https://doi.org/10.1641/0006-3568(2004)054[0066:tvomcf]2.0.co;2
- Data issues in the life sciences.ZooKeys150:15. https://doi.org/10.3897/zookeys.150.1766
- SDD and the key to life, TDWG 2005.TDWG,1pp. URL: http://www.tdwg.org/2005meet/TDWG2005_Abstract_71.htm
- An appraisal of megascience platforms for biodiversity information.MycoKeys5:45‑63. https://doi.org/10.3897/mycokeys.5.4302
- Flora of the Southern and Mid-Atlantic States. Working Draft of 30 November 2012.Online Publication,1225pp. URL: http://www.herbarium.unc.edu/FloraArchives/WeakleyFlora_2012-Nov.pdf
- Small Pieces Loosely Joined: A Unifying Theory of the Web.Basic Books,New York,240pp. URL: http://www.smallpieces.com/index.php
- Mapping the biosphere: exploring species to understand the origin, organization and sustainability of biodiversity.Systematics and Biodiversity10:1‑20. https://doi.org/10.1080/14772000.2012.665095
- Darwin Core: An Evolving Community-Developed Biodiversity Data Standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
- The encyclopedia of life.Trends in Ecology & Evolution18(2):77‑80. https://doi.org/10.1016/s0169-5347(02)00040-x