|
Biodiversity Data Journal :
Research Article
|
|
Corresponding author: Sarah J. Bourlat (S.Bourlat@leibniz-zfmk.de)
Academic editor: Urmas Kõljalg
Received: 15 Mar 2017 | Accepted: 06 Jun 2017 | Published: 08 Jun 2017
© 2017 Quiterie Haenel, Oleksandr Holovachov, Ulf Jondelius, Per Sundberg, Sarah Bourlat
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Haenel Q, Holovachov O, Jondelius U, Sundberg P, Bourlat S (2017) NGS-based biodiversity and community structure analysis of meiofaunal eukaryotes in shell sand from Hållö island, Smögen, and soft mud from Gullmarn Fjord, Sweden. Biodiversity Data Journal 5: e12731. https://doi.org/10.3897/BDJ.5.e12731
|
|
Abstract
Aim: The aim of this study was to assess the biodiversity and community structure of Swedish meiofaunal eukaryotes using metabarcoding. To validate the reliability of the metabarcoding approach, we compare the taxonomic resolution obtained using the mitochondrial cytochrome oxidase 1 (COI) ‘mini-barcode’ and nuclear 18S small ribosomal subunit (18S) V1-V2 region, with traditional morphology-based identification of Xenacoelomorpha and Nematoda.
Location: 30 samples were analysed from two ecologically distinct locations along the west coast of Sweden. 18 replicate samples of coarse shell sand were collected along the north-eastern side of Hållö island near Smögen, while 12 replicate samples of soft mud were collected in the Gullmarn Fjord near Lysekil.
Methods: Meiofauna was extracted using flotation and siphoning methods. Both COI and 18S regions were amplified from total DNA samples using Metazoan specific primers and subsequently sequenced using Illumina MiSeq, producing in total 24 132 875 paired-end reads of 300 bp in length, of which 15 883 274 COI reads and 8 249 601 18S reads. These were quality filtered resulting in 7 954 017 COI sequences and 890 370 18S sequences, clustered into 2805 and 1472 representative OTUs respectively, yielding 190 metazoan OTUs for COI and 121 metazoan OTUs for 18S using a 97% sequence similarity threshold.
Results: The Metazoan fraction represents 7% of the total dataset for COI (190 OTUs) and 8% of sequences for 18S (121 OTUs). Annelida (30% of COI metazoan OTUs and 23.97% of 18S metazoan OTUs) and Arthropoda (27.37% of COI metazoan OTUs and 11.57% of 18S metazoan OTUs), were the most OTU rich phyla identified in all samples combined. As well as Annelida and Arthropoda, other OTU rich phyla represented in our samples include Mollusca, Platyhelminthes and Nematoda. In total, 213 COI OTUs and 243 18S OTUs were identified to species using a 97% sequence similarity threshold, revealing some non-native species and highlighting the potential of metabarcoding for biological recording. Taxonomic community composition shows as expected clear differentiation between the two habitat types (soft mud versus coarse shell sand), and diversity observed varies according to choice of meiofaunal sampling method and primer pair used.
Keywords
Meiofaunal biodiversity, community structure, Illumina Mi-Seq, Metabarcoding, COI, 18S
Introduction
Microscopic interstitial marine organisms, also termed ‘meiofauna’, are often defined as animals that pass a 1mm mesh but are retained on a 45 µm sieve (
Methodological comparison of benthic and pelagic metabarcoding studies of marine fauna published to date
|
Authors |
Sample type |
Sample extraction method |
Sequencing platform |
Marker |
Marker size (bp) |
Chimera screening |
OTU clustering method and threshold |
Database |
|
|
Coral reef fish gut contents |
Dissection of fish gut |
Roche 454 GS FLX |
COI |
313 |
UCHIME |
CROP 92-94% |
Moorea Biocode Database, GenBank |
|
|
Autonomous reef monitoring structures |
4 fractions (Sessile, 2mm, 500μm, 106μm) |
Ion Torrent |
COI |
313 |
|
|
BOLD, GenBank |
|
|
Zooplankton from 50m to the surface |
200μm mesh WP2 plankton net |
Roche 454 GS FLX |
18S (V1-V2 regions) |
450 |
ChimeraSlayer (QIIME 1.3.0) |
UCLUST 97% (QIIME 1.3.0) |
Silva 108, GenBank |
|
|
Plankton |
3 fractions (5-20μm, 20-180μm, 180-2000μm) |
Paired-end Illumina Genome Analyser IIx system |
18S (V9 region) |
|
USEARCH |
|
V9_PR2, V9 rDNA, Protistan Ribosomal Reference Database |
|
|
Marine benthic meiofauna |
Decanting 45μm sieve Ludox |
Roche 454 GS FLX |
18S (V1-V2 regions) |
364 (250-500) |
OCTOPUS |
OCTOPUS 96% |
GenBank |
|
|
Marine benthic meiofauna |
Decanting 45μm sieve Ludox |
Roche 454 GS FLX |
18S (V1-V2 regions) |
450 |
Amplicon-Noise |
Amplicon-Noise 99% and 96% |
GenBank |
|
|
Marine benthic meiofauna |
Directly from sediment, elutriated on 45μm sieve |
Paired-end 100 bp reads Illumina HiSeq |
18S (V9 region) |
87-187 [13] |
USEARCH 6.1. (QIIME 1.8) |
UPARSE 97% UCLUST and USEARCH (QIIME 1.8) |
Silva 111 |
|
|
Benthic meiofauna from seagrass meadows |
2mm sieve, 1mm sieve, 0.5mm sieve |
Roche 454 GS FLX |
COI 18S |
450 710 |
USEARCH 6.1 (QIIIME 1.7) |
UCLUST de novo (QIIME 1.7) |
GenBank Silva 115 |
|
This study |
Meiofauna from coarse shell sand and muddy benthic sediment |
Siphoning 125μm, flotation (MgCl2) 125μm, flotation (H2O) 45μm/70μm |
Paired-end Illumina Mi-Seq |
COI 18S (V1-V2 regions) |
313 364 |
UCHIME (part of USEARCH 6.1.) (QIIME 1.9.1) |
CROP COI: 92-94% 18S: 95-97% |
BOLD, SweBol and own databases for Nemertea, Acoela, Oligochaeta), Genbank Silva 111 |
In this study we used samples from muddy and sandy marine sediments to examine how results of metabarcoding based surveys of meiofaunal communities are impacted by three different meiofaunal extraction methods and three different primer pairs for COI and 18S. In order to validate the reliability of the metabarcoding approach, we compare the results obtained with traditional morphology-based taxonomic assignment for two test groups, Xenacoelomorpha and Nematoda, the latter previously shown to be the dominant taxon in meiofaunal communities in terms of number of OTUs (
Materials and Methods
Sampling
Samples were collected in two ecologically distinct locations along the west coast of Sweden in August 2014.
Hållö island samples: Coarse shell sand was sampled by dredging at 7-8m depth along the north-eastern side of Hållö island near Smögen, Sotenäs municipality, Västra Götalands county (
Gullmarn Fjord samples: Soft mud was collected using a Waren dredge at 53 m depth in the Gullmarn Fjord near Lysekil, Lysekil municipality, Västra Götalands county (
Meiofaunal extraction
Hållö island. Hållö island samples were extracted in the lab using two different variations of the flotation (decanting and sieving) technique.
Flotation (freshwater): Freshwater was used to induce an osmotic shock in meiofaunal organisms and force them to detach from heavy sediment particles. 200 mL of sediment were placed in a large volume of fresh water and thoroughly mixed to suspend meiofauna and lighter sediment particles. The supernatant was sieved through a 1000 µm sieve to separate the macrofaunal fraction, which was then discarded. The filtered sample was sieved again through a 45 µm sieve to collect meiofauna and discard fine organic particles. This procedure was repeated three times. Meiofauna was then rinsed with seawater from the sieve into large falcon tubes. Twelve sediment samples were processed, ten of them were fixed immediately in 96% ethanol for molecular analysis and stored at -20°C. The other two samples were first screened for live representatives of Xenacoelomorpha, and later preserved in 4% formaldehyde for morphology-based identification of nematodes.
Flotation (MgCl2 solution): A 7.2% solution of MgCl2 was used to anesthetize meiofauna. As above, twelve samples were processed in total, ten of them were decanted through 125 µm sieve and fixed immediately in 96% ethanol for molecular analysis and stored at -20°C, while two samples were decanted through a 125 µm sieve which was subsequently placed in a petri dish with seawater. After 30 minutes, the petri dish as well as the inside of the sieve were searched for Xenacoelomorpha using a stereo microscope. Afterwards they were preserved in 4% formaldehyde for morphology-based identification of nematodes.
Gullmarn Fjord. Meiofauna was extracted from the Gullmarn Fjord samples using two different methods: flotation and siphoning.
Flotation (freshwater): Freshwater was used to induce an osmotic shock in meiofaunal organisms. 2.4 L of sediment were placed in a large volume of freshwater, thoroughly mixed to suspend meiofauna and lighter sediment particles. The supernatant was sieved through a 1000 µm sieve in order to separate macrofauna, which was then discarded. The filtered sample was then sieved three times through a 70µm sieve to collect meiofauna and discard fine organic particles. Meiofauna was then rinsed with seawater from the sieve into a large container and equally divided between 12 falcon tubes. Six samples were fixed in 96% ethanol for molecular analysis and stored at -20°C. Six samples were screened for live representatives of Xenacoelomorpha, and preserved in 4% formaldehyde for morphology-based identification of nematodes.
Siphoning: A total volume of 12 L of sediment was processed as follows: an approximately 5 cm thick layer of mud was placed in a container and covered with 20 cm of seawater. The sediment was allowed to settle for 20 hours. Half of the sediment area was then siphoned through a 125 µm sieve, the residue in the sieve was immediately fixed in 96% ethanol, large macrofauna was manually removed, and the entire volume was split equally into six samples and placed at -20°C for subsequent molecular analysis. The remaining half of the area was similarly siphoned through a 125 µm sieve, the sieve contents were stored in sea water, large macrofauna manually removed, the entire volume split into six samples, which were screened for live representatives of Xenacoelomorpha, and preserved in 4% formaldehyde for morphology-based identification of nematodes.
Morphology-based identification
Xenacoelomorpha. Four samples from Hållö and 12 samples from Gullmarn Fjord were used for morphology-based assessment of the diversity of Xenacoelomorpha. All samples were stored in seawater and searched for Xenacoelomorpha with a stereo microscope. All specimens found were immediately identified to the lowest taxonomic rank possible using a compound microscope equipped with DIC.
Nematoda. Two samples from each location/extraction method were used to assess nematode diversity using morphology-based identification. Samples from Hållö (flotation with fresh water and MgCl2) and Gullmarn Fjord (siphoning) were processed whole and samples from Gullmarn Fjord extracted using flotation with fresh water were subsampled by taking 1/10 of the entire sample. Formaldehyde–preserved samples were transferred to glycerin using Seinhorst’s rapid method as modified by
DNA extraction, library preparation and sequencing
DNA extraction. 30 samples were processed for total DNA extraction, twelve from the Gullmarn Fjord and eighteen from Hållö island, using 10g of sediment and the PowerMax® Soil DNA Isolation Kit (MO BIO Laboratories), according to manufacturer’s instructions.
Primer design. Illumina MiSeq reagent v3. produces paired-end reads of 300bp in length, allowing a maximum marker length of 500bp when taking into account a 50 bp overlap. Universal COI primers available for the Metazoa amplify a 658bp region (
Accordingly, primers amplifiying a 313 bp fragment of the mitochondrial cytochrome oxidase 1 (COI) gene were used, as described in
For the 18S region, Illumina overhang adapter sequences were appended to the primers from Fonseca et al. (SSU_FO4-SSU_R22;
Sequence overlap in the paired-end reads was calculated in Geneious
All primer sequences used are shown in Table
|
Marker |
Primer name |
Illumina adapter overhang (regular font), with primer sequence (in bold) |
|
COI Leray |
mlCOIintF |
5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGWACWGGWTGAACW GTWTAYCCYCC-3’ |
|
dgHCO2198 |
5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTAAACTTCAGGGTGAC CAAARAAYCA-3’ |
|
|
COI Lobo |
mlCOIintF |
5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGWACWGGWTGAACW GTWTAYCCYCC-3’ |
|
LoboR1 |
5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTAAACYTCWGGRTGW CCRAARAAYCA-3’ |
|
|
18S |
SSU_FO4 |
5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGCTTGTCTCAAAGATTA AGCC-3’ |
|
SSU_R22 |
5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGCCTGCTGCCTTCCTT GGA-3’ |
Illumina MiSeq library preparation using fusion primers. For Illumina MiSeq library preparation, we used a dual PCR amplification method as described in
Amplicon PCR. PCR amplifications of the COI and 18S regions were set up as follows. For a 50µl reaction volume, we used 5µl Pfu polymerase buffer (10x), 1µl dNTP mix (final concentration of each dNTP 200µM), 0.5 µl of each primer at 50 pm/µl, 2 µl DNA template (~10 ng), 0.5µl Pfu DNA polymerase (Promega) and 40.5µl of nuclease free water. Each DNA sample was amplified with the 3 primer pairs described above (COI Leray, COI Lobo and 18S). PCR cycling conditions were 2 min at 95°C (1 cycle); 1 min at 95°C, 45 s at 57°C, 2 min at 72°C (35 cycles); 10 min at 72°C (1 cycle). The PCR was checked on a 2% agarose gel. 20µl of each PCR reaction were then purified with Agencourt® AMPure® XP paramagnetic beads (Beckman Coulter), allowing size selection of PCR fragments by using different PCR product to bead ratios (
Index PCR. For dual indexing we used the Nextera XT index kit (96 indices, 384 samples, Illumina) according manufacturers’ instructions. Dual indexing allows an increase in the multiplex level of sequencing per lane, so that more samples can be sequenced on the same flow cell (
Sequencing. The pooled libraries were sequenced three times independently using Illumina MiSeq Reagent Kit v3, producing in total 24 132 875 paired-end reads of 300 bp in length, of which 15 883 274 COI reads and 8 249 601 18S reads (Table
Bioinformatic data processing and analysis
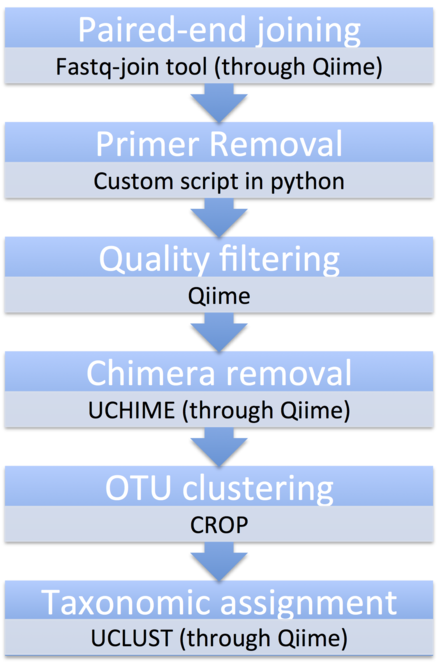
Most analytical steps were performed using Qiime (Quantitative Insight Into Microbial Ecology) version 1.9.1 (
Paired-end joining
Demultiplexed MiSeq paired-end reads were joined using the Qiime script multiple_join_paired_ends.py using the fastq-join tool (https://code.google.com/p/ea-utils/wiki/FastqJoin). Data from three sequencing runs were merged producing a total of 24 132 875 raw paired-end reads, 15 883 274 reads for the COI dataset and 8 249 601 reads for the 18S dataset (Table
|
Marker / Step |
Raw data |
Paired-end joining |
Primer trimming |
Quality filtering |
Chimera removal |
|
COI |
15 883 274 |
10 412 096 |
8 099 507 |
7 976 649 |
7 954 017 |
|
18S |
8 249 601 |
2 131 102 |
1 071 871 |
1 015 874 |
890 370 |
|
Total |
24 132 875 |
12 543 198 |
9 171 378 |
8 992 523 |
8 844 387 |
Primer trimming and quality filtering
Dual indexes and Illumina overhangs were removed by the sequencing platform. COI and 18s primer sequences were removed using a custom python script designed for this study (https://github.com/Quiterie90/Primer_Removal). The script retains and trims reads that have the exact sequence of the forward and reverse primers at the beginning and at the end of the reads respectively, while other reads not meeting these criteria are discarded. The script takes into account the presence of ambiguous bases in the primer sequence (such as W, R, S, Y, M, K, H, D, B and V). In the case that an unassigned base (N) is found in the primer sequence, the read is also discarded. The primer-trimming step resulted in 9 171 378 reads remaining corresponding to a 27% loss. As the script is quite stringent, it quality filters reads by removing incomplete reads or chimeras. At this step 1 071 871 reads remained after trimming for the 18S dataset corresponding to a 50% loss and 8 099 507 reads remained after trimming for the COI dataset corresponding to a 22% loss. A quality filtering step was then carried out using the Qiime script multiple_split_ libraries_fastq.py to remove reads with a Q Score inferior to 30 (corresponding to a base call accuracy of at least 99,9%). A total of 2% of sequences were lost after the quality-filtering step leading to 8 992 523 reads remaining. 5% of the reads were lost in the 18S dataset corresponding to a final 1 015 874 reads and 1,5% of the reads were lost in the COI dataset corresponding to a final 7 976 649 reads.
Chimera removal and OTU clustering
Chimeric reads were removed with UCHIME (
For clustering sequences into Operational Taxonomic Units (OTUs) we used CROP, a Bayesian clustering algorithm that delineates OTUs based on the natural distribution of the data, using a Gaussian mixture model (
Parameters used in CROP for the analysis were as follows:
CROP -i <input_CO1.fasta> -b 160 000 -z 470 -l 3 -u 4 -o <output_CO1>
CROP -i <input_18S.fasta> -b 18 000 -z 470 -l 1.5-u 2.5 -o <output_18S
The 7 954 017 COI sequences and the 890 370 18S sequences were clustered into 2805 and 1472 representative OTUs respectively, 213 of which were identified to species for COI and 243 of which were identified to species for 18S, using a 97% sequence similarity threshold (Table
Number of OTUs and percentage per phylum for COI and 18S for the metazoan fraction. Based on a 97% similarity threshold.
|
Phylum |
COI |
18S |
||
|
OTUs |
Percentage |
OTUs |
Percentage |
|
|
Annelida |
57 |
30.00 |
29 |
23.97 |
|
Arthropoda |
52 |
27.37 |
14 |
11.57 |
|
Bryozoa |
5 |
2.63 |
3 |
2.48 |
|
Cephalorhyncha |
0 |
0.00 |
1 |
0.83 |
|
Chaetognatha |
1 |
0.53 |
0 |
0.00 |
|
Chordata |
12 |
6.32 |
7 |
5.79 |
|
Cnidaria |
8 |
4.21 |
4 |
3.31 |
|
Echinodermata |
13 |
6.84 |
5 |
4.13 |
|
Gastrotricha |
1 |
0.53 |
9 |
7.44 |
|
Gnathostomulida |
1 |
0.53 |
0 |
0.00 |
|
Mollusca |
26 |
13.68 |
6 |
4.96 |
|
Nematoda |
0 |
0.00 |
10 |
8.26 |
|
Nemertea |
3 |
1.58 |
6 |
4.96 |
|
Platyhelminthes |
0 |
0.00 |
13 |
10.74 |
|
Phoronida |
1 |
0.53 |
0 |
0.00 |
|
Porifera |
2 |
1.05 |
3 |
2.48 |
|
Priapulida |
1 |
0.53 |
0 |
0.00 |
|
Rotifera |
2 |
1.05 |
0 |
0.00 |
|
Sipuncula |
1 |
0.53 |
1 |
0.83 |
|
Tardigrada |
0 |
0.00 |
1 |
0.83 |
|
Xenacoelomorpha |
4 |
2.11 |
9 |
7.44 |
|
Total OTUs Metazoa |
190 |
100 |
121 |
100 |
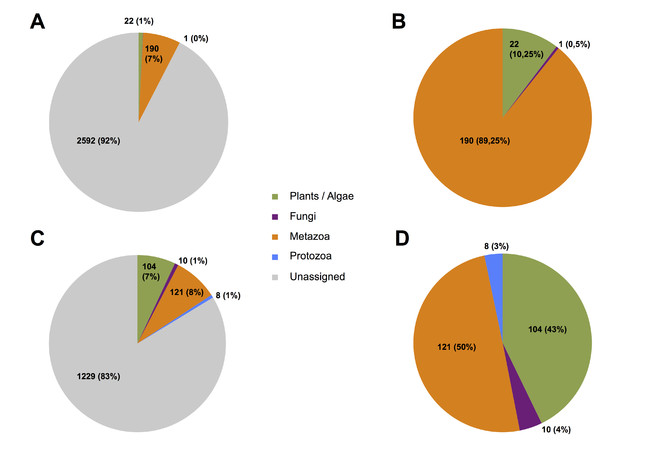
Taxonomic composition overview at species level based on a 97% sequence similarity threshold. A) Percentages and counts of OTUs for the COI gene with unassigned OTUs. B) Percentages and counts of OTUs for the COI gene without unassigned OTUs. C) Percentages and counts of OTUs for the 18S gene with unassigned OTUs. D) Percentages and counts of OTUs for the 18S gene without unassigned OTUs.
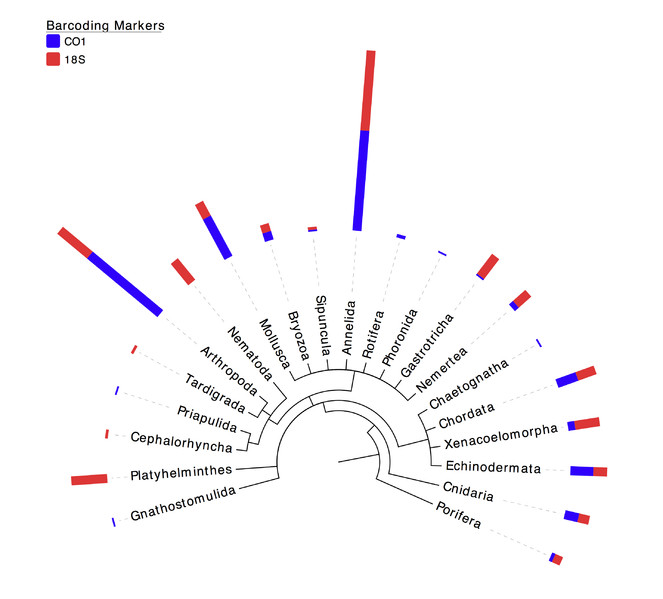
Percentages of metazoan phyla uncovered in the samples using COI and 18S molecular surveys. Blue bars correspond to the cumulated frequencies of OTUs assigned to a specific phylum using the COI gene and red bars correspond to the cumulated frequencies of OTUs assigned to a specific phylum using the 18S gene. Taxonomic assignment is based on a 97% sequence similarity threshold.
Taxonomic assignment
As Qiime is normally used for metagenomic analyses of prokaryotes, default databases are not suited for taxonomic assignment of Metazoa. Custom databases consisting in a taxonomy file associated with a reference sequence file can be created, or alternatively, a preformatted database such as the Silva database (http://www.arb-silva.de/no_cache/download/archive/qiime/) can be used. For the COI region, a custom database of 1 947 954 sequences was created consisting of the BOLD database (http://www.boldsystems.org/ downloaded on October 8 2015), combined with own reference databases of Nemertea, Xenacoelomorpha and Oligochaeta and barcodes of Swedish Echinodermata, Mollusca, Cnidaria and Arthropoda from the Swedish Barcode of Life database (SweBol). For the 18S rRNA region, a custom database of 732 419 reference sequences was created using the Silva database release 111 (http://www.arb-silva.de/no_cache/download/archive/qiime/) and own barcodes for Acoela and Oligochaeta. Corresponding tab-delimited taxonomy files were created including a sequence ID and taxonomic lineage information (Phylum, Class, Order, Family, Genus and Species) derived from BOLD, Swebol, Silva and WoRMS (http://www.marinespecies.org/).
Taxonomic assignments were carried out using both 80% and 97% sequence similarity thresholds, to obtain identifications at phylum and species levels respectively (
Note on the taxonomic assignement of Nematodes: The output from the Qiime analysis included 145 18S OTUs assigned to the phylum Nematoda. Three of them (HE1.SSU866120, HE6.SSU382930 and HF6.SSU331569) Suppl. material 1 were incorrectly placed among the nematodes due to errors in the reference database they derived from – they group among Arthropod taxa by the Megablast search and were excluded for that reason. Another OTU (TS6.SSU559982) is placed among Phoronida by the Megablast search and was also excluded. Two more sequences that were assigned to Nematoda appear to have long insertions within conserved regions (HE6.SSU358113 and TF5.SSU411806). Both of them were found only in one sample each, further supporting the idea that they are derived from erroneous amplification product, and were removed from any further analysis.
Invasive Alien Species (IAS) were detected in our samples by comparing our species list (Suppl. material
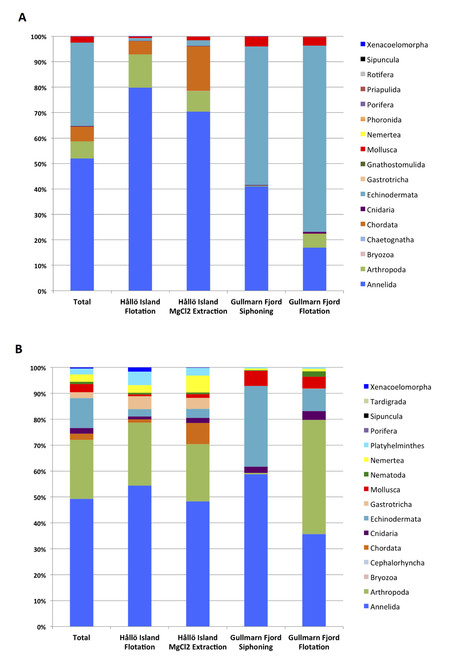
Taxonomic composition bar plots (Fig.
Community composition per phylum in Hållö island and Gullmarn fjord samples, according to extraction method (MgCl2, H2O, Siphoning). A) For the COI gene. B) For the 18S gene. The vertical axis corresponds to percentage of OTUs. Taxonomic assignment is based on a 97% similarity threshold. The bar plots take into account number of reads for each OTU.
Diversity analyses
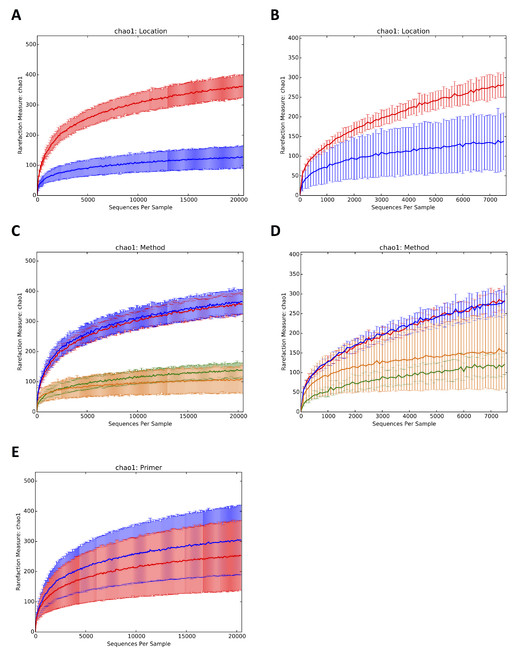
Alpha and beta diversity analyses were carried out with and without unassigned OTUs for both COI and 18S datasets. Unassigned OTUs were removed using the Qiime script filter_otus_from_otu_table.py. Alpha diversity (species richness) was calculated using the nonparametric Chao1 index using rarefied datasets to correct bias in species number due to unequal sample size. One of the samples in the COI dataset was removed prior to rarefaction analysis due to low sequence number (1122 sequences including unassigned OTUs and 280 sequences excluding unassigned OTUs at 97% sequence similarity) using the Qiime script filter_sample_from_otu_table.py. Rarefaction, alpha diversity calculation and generation of plots were performed using the Qiime scripts i) multiple_rarefactions.py, ii) alpha_diversity.py, iii) collate_alpha.py and iv) make_rarefaction_plots.py. Rarefaction was done to a depth corresponding to the total number of sequences in the smallest dataset (20405 sequences including unassigned OTUs and 5442 sequences excluding unassigned OTUs at 97% sequence similarity for COI, and 7561 sequences including unassigned OTUs and 5399 sequences excluding unassigned OTUs at 97% sequence similarity for 18S). Alpha diversities were compared between locations and extraction methods for both datasets and COI primer sets using the Qiime script compare_alpha_diversity.py. The script performs Monte-Carlo permutations to determine p-values.
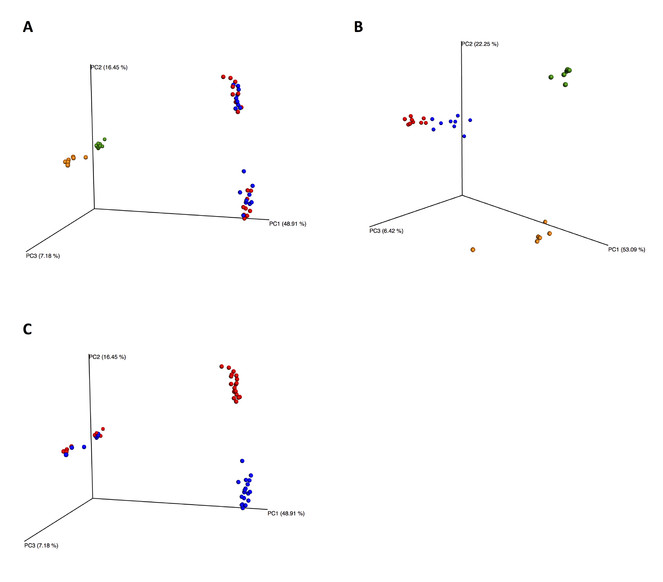
Beta diversity was calculated using the abundance-based Bray-Curtis index for both COI and 18S datasets. The Qiime script beta_diversity_through_plots.py was used to compute beta diversity distance matrices from the rarefied samples and generate Principal Coordinate Analysis (PCoA) plots. Beta diversity was compared according to location, extraction method and primer pair both with and without the unassigned OTUs using the Qiime script compare_categories.py. The script uses R and the vegan and ape libraries to compute statistical tests. We performed ANOSIM (ANalysis Of SIMilarity) tests, which are nonparametric, through 999 permutations. This method tests whether two or more groups of samples are significantly different by taking as null hypothesis that there is no difference between the two or more groups studied.
Alpha and beta diversities were calculated including and excluding the unassigned OTUs and results obtained were similar. Here we present plots including the unassigned OTUs (Figs
Alpha diversity rarefaction plots for COI and 18S datasets including unassigned OTUs. According to location for COI (A) 18S (B). Hållö Island (HI) in red, Gullmarn Fjord (GF) in blue. According to extraction method for COI (C) 18S (D). HI flotation in red, HI MgCl2 in blue, GF flotation in yellow, GF siphoning in green. According to primer pair for COI (E). CO1 Leray primer in red, COI Lobo primer in blue.
Beta diversity PCoA plots for COI and 18S datasets including unassigned OTUs. According to extraction method for COI (A) 18S (B) HI flotation in red, HI MgCl2 in blue, GF flotation in yellow and GF siphoning in green. According to primer for COI (C) COI Leray primer in red, COI Lobo primer in blue
Data resources
The data underpinning the analysis reported in this paper are deposited at the GenBank SRA under project number PRJNA388326 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA388326).
Results and discussion
Phylum-level community composition of meiofaunal samples from the Swedish west coast
Illumina MiSeq produced at total of 24 132 875 raw reads, of which 15 883 274 COI reads and 8 249 601 18S reads. These were quality filtered (see methods section for details) resulting in 7 954 017 COI sequences and 890 370 18S sequences. These were clustered into 2805 and 1472 representative OTUs respectively, yielding 190 metazoan OTUs for COI and 121 metazoan OTUs for 18S at 97% sequence similarity (see methods, Table
Taxonomic assignment of OTUs at a 97% similarity threshold shows community composition of the samples at the phylum level (Fig.
For the 18S dataset, 121 of 1472 OTUs (8%) were assigned to Metazoa, 104 (7%) to plants and algae, 10 (1%) to Fungi, and 8 (1%) to Protozoa. 1229 OTUs remained unassigned, corresponding to 83% of all 18S OTUs.
The large numbers of unassigned OTUs reflect the incompleteness of the databases used for COI and 18S. When unassigned OTUs are disregarded, differences between the taxonomic ocverage of the markers can be observed (Fig.
Of all OTUs classified as Metazoa, a detailed breakdown per phylum is presented in Table
As well as Annelida and Arthropoda, other phyla represented by a high number of OTUs in our samples include Mollusca (13.68% of COI metazoan OTUs and 4.96% of 18S metazoan OTUs), Platyhelminthes (10,74% of 18S metazoan OTUs and 0% of CO1 metazoan OTUs) and Nematoda (8.26% of 18S metazoan OTUs and 0% of CO1 metazoan OTUs) (Table
Meiofaunal community composition differs according to location
Taxonomic community composition at both locations surveyed is illustrated in Fig.
In Fig.
In coarse shell sand in shallow areas, such as in the Hållö island samples, Annelida and Arthropoda are represented by higher numbers of reads, followed by Chordata (cephalohordata such as Branchiostoma sp., ascidians and various fish species such as Gobius sp., Ctenolabrus rupestris, Solea solea) with in addition a larger diversity of small taxa such as Bryozoa, Gnathosthomulida, Gastrotricha, Tardigrada, Rotifera, Sipuncula and Phoronida, reflecting the high diversity of insterstitial taxa found in sandy sediments.
Sample diversity and composition analyses
A greater number of phyla were uncovered in the Hållö Island samples than in the Gullmarn Fjord samples (Fig.
Nonparametric t-test results with 999 Monte-Carlo permutations for both datasets with and without unassigned OTUs (97% taxonomic assignment)
|
|
COI dataset |
18S dataset |
||||||
|
|
Excluding unassigned OTUs |
Including Unassigned OTUs |
Excluding unassigned OTUs |
Including Unassigned OTUs |
||||
|
|
Test value |
P-value |
Test value |
P-value |
Test value |
P-value |
Test value |
P-value |
|
Location |
|
|
|
|
|
|
|
|
|
HI vs. GF |
-14.453 |
0.001 |
-21.455 |
0.001 |
-6.929 |
0.001 |
-7.170 |
0.001 |
|
Method |
|
|
|
|
|
|
|
|
|
HI H2O vs. HI MgCl2 |
-0.437 |
1.0 |
-0.691 |
1.0 |
-0.906 |
1.0 |
-0.174 |
1.0 |
|
GF flotation vs. GF siphoning |
1.567 |
0.792 |
1.546 |
0.99 |
-1.427 |
1.0 |
-0.744 |
1.0 |
|
Primer |
|
|
|
|
|
|
|
|
|
COI Leray vs. COI Lobo |
-0.508 |
0.596 |
-1.614 |
0.111 |
- |
- |
- |
- |
ANOSIM test results (999 permutations) for both COI and 18S datasets with and without unassigned OTUs (97% taxonomic assignment)
|
|
COI dataset |
18S dataset |
||||||
|
Ho: Sample composition differs according to |
Excluding unassigned OTUs |
Including unassigned OTUs |
Excluding unassigned OTUs |
Including unassigned OTUs |
||||
|
|
R-value |
P-value |
R-value |
P-value |
R-value |
P-value |
R-value |
P-value |
|
Location |
0.976 |
0.001 |
1.0 |
0.001 |
0.935 |
0.001 |
0.929 |
0.001 |
|
Method |
0.660 |
0.001 |
0.738 |
0.001 |
0.889 |
0.001 |
0.895 |
0.001 |
|
Primer |
0.200 |
0.003 |
0.218 |
0.001 |
- |
- |
- |
- |
Molecular identifications to species level
Using a sequence similarity search at 97% similarity allowed us to identify 213 COI OTUs and 243 18S OTUs to species level (Table
Metazoa identified to species level using 97% sequence similarity (HI: Hållö island, GF: Gullmarn Fjord)
| COI | |||||||
| OTU ID | Nb of reads | Phylum | Class | Order | Species | HI | GF |
| HE6.Lobo_7972794 | 3 | Annelida | Clitellata | Haplotaxida | Adelodrilus pusillus | + | - |
| HE1.Lobo_933012 | 14954 | Annelida | Clitellata | Haplotaxida | Grania postclitellochaeta | + | + |
| HF8.Lobo_5239705 | 241 | Annelida | Clitellata | Haplotaxida | Grania variochaeta | + | + |
| HF4.Lobo_97092 | 29391 | Annelida | Clitellata | Haplotaxida | Tubificoides benedii | + | + |
| HF5.Lobo_3297996 | 1 | Annelida | Clitellata | Haplotaxida | Tubificoides kozloffi | + | - |
| TS1.Leray_545620 | 7370 | Annelida | Polychaeta | Amphinomida | Paramphinome jeffreysii | - | + |
| HF1.Lobo_4996219 | 4596 | Annelida | Polychaeta | Canalipalpata | Polygordius appendiculatus | + | + |
| TF6.Lobo_5247622 | 9030 | Annelida | Polychaeta | Capitellida | - | + | |
| TS1.Lobo_4669404 | 5 | Annelida | Polychaeta | Capitellida | - | + | |
| TF5.Lobo_6394093 | 2 | Annelida | Polychaeta | Capitellida | - | + | |
| TS3.Leray_6813257 | 1852 | Annelida | Polychaeta | Eunicida | - | + | |
| HF5.Leray_4035802 | 1 | Annelida | Polychaeta | Eunicida | Ophryotrocha maculata | + | - |
| TS2.Leray_4445240 | 8815 | Annelida | Polychaeta | Eunicida | Parougia eliasoni | + | + |
| TF3.Leray_6645504 | 5196 | Annelida | Polychaeta | Opheliida | + | + | |
| TS5.Lobo_6031643 | 5089 | Annelida | Polychaeta | Opheliida | + | + | |
| HF9.Lobo_7587930 | 1 | Annelida | Polychaeta | Opheliida | + | - | |
| HE8.Leray_7284535 | 2 | Annelida | Polychaeta | Phyllodocida | + | - | |
| TS5.Leray_1557252 | 88 | Annelida | Polychaeta | Phyllodocida | - | + | |
| TS3.Leray_6744085 | 1 | Annelida | Polychaeta | Phyllodocida | - | + | |
| TS3.Leray_6805306 | 2 | Annelida | Polychaeta | Phyllodocida | Aphrodita aculeata | - | + |
| TS3.Lobo_1308935 | 4213 | Annelida | Polychaeta | Phyllodocida | Eumida ockelmanni | + | + |
| HE6.Leray_2958692 | 69642 | Annelida | Polychaeta | Phyllodocida | Glycera alba | + | + |
| HF7.Leray_1672792 | 69 | Annelida | Polychaeta | Phyllodocida | Glycinde nordmanni | + | + |
| TF5.Leray_2872180 | 7754 | Annelida | Polychaeta | Phyllodocida | Gyptis mackiei | - | + |
| HF1.Lobo_5059232 | 13 | Annelida | Polychaeta | Phyllodocida | Gyptis propinqua | + | - |
| HF9.Lobo_7695035 | 1 | Annelida | Polychaeta | Phyllodocida | Lepidonotus squamatus | + | - |
| HE6.Lobo_7972042 | 2 | Annelida | Polychaeta | Phyllodocida | Myrianida edwarsi | + | - |
| HF9.Lobo_7688887 | 3 | Annelida | Polychaeta | Phyllodocida | Nereimyra punctata | + | - |
| HF2.Lobo_2136301 | 178929 | Annelida | Polychaeta | Phyllodocida | Pisione remota | + | + |
| HE3.Leray_364663 | 59407 | Annelida | Polychaeta | Phyllodocida | Platynereis dumerilli | + | + |
| TS4.Leray_7471107 | 1 | Annelida | Polychaeta | Phyllodocida | Sige fusigera | - | + |
| HE5.Lobo_493462 | 571790 | Annelida | Polychaeta | + | + | ||
| TS2.Lobo_6962270 | 4595 | Annelida | Polychaeta | Sabellida | Galathowenia oculata | + | + |
| TS2.Leray_4491798 | 316559 | Annelida | Polychaeta | Spionida | + | + | |
| TS4.Lobo_1502925 | 195999 | Annelida | Polychaeta | Spionida | + | + | |
| HF9.Lobo_7588557 | 891 | Annelida | Polychaeta | Spionida | + | - | |
| TS6.Leray_5665274 | 936 | Annelida | Polychaeta | Spionida | - | + | |
| TF1.Lobo_2668551 | 874 | Annelida | Polychaeta | Spionida | - | + | |
| HE4.Leray_3067470 | 3 | Annelida | Polychaeta | Spionida | Chaetopterus sarsi | + | - |
| HF1.Lobo_4965916 | 1 | Annelida | Polychaeta | Spionida | Malacoceros fuliginosus | + | - |
| HF9.Leray_4404528 | 1 | Annelida | Polychaeta | Spionida | Polydora cornuta | + | - |
| HF5.Lobo_3178682 | 2894 | Annelida | Polychaeta | Spionida | Spiophanes bombyx | + | + |
| TF1.Leray_2314881 | 29235 | Annelida | Polychaeta | Terebellida | + | + | |
| TF1.Lobo_2832834 | 9348 | Annelida | Polychaeta | Terebellida | + | + | |
| TS1.Leray_614419 | 788 | Annelida | Polychaeta | Terebellida | + | + | |
| HE8.Lobo_858951 | 1 | Annelida | Polychaeta | Terebellida | + | - | |
| TS2.Lobo_6889557 | 184 | Annelida | Polychaeta | Terebellida | - | + | |
| TS6.Lobo_255019 | 3 | Annelida | Polychaeta | Terebellida | - | + | |
| TS2.Lobo_6860909 | 1 | Annelida | Polychaeta | Terebellida | - | + | |
| TS5.Leray_1638640 | 1 | Annelida | Polychaeta | Terebellida | - | + | |
| TF1.Lobo_2848745 | 1305 | Annelida | Polychaeta | Terebellida | Amphictene auricoma | + | + |
| TS3.Leray_6729893 | 1 | Annelida | Polychaeta | Terebellida | Brada villosa | - | + |
| HF4.Lobo_96799 | 102 | Annelida | Polychaeta | Terebellida | Cirratulus cirratus | + | - |
| HF2.Lobo_2052205 | 285 | Annelida | Polychaeta | Terebellida | Dodecaceria concharum | + | - |
| TS5.Leray_1638834 | 102 | Annelida | Polychaeta | Terebellida | Lagis koreni | + | + |
| HE9.Lobo_2191024 | 8 | Annelida | Polychaeta | Terebellida | Macrochaeta clavicornis | + | + |
| TF1.Leray_2475372 | 6353 | Annelida | Polychaeta | Terebellida | Sosane wahrbergi | + | + |
| HE1.Lobo_982378 | 38 | Arthropoda | Branchiopoda | Diplostraca | Evadne nordmanni | + | - |
| TF5.Lobo_6391642 | 10097 | Arthropoda | Branchiopoda | Diplostraca | Penilia avirostris | + | + |
| HF9.Lobo_7623741 | 1 | Arthropoda | Branchiopoda | Diplostraca | Pleopis polyphemoides | + | - |
| TS4.Leray_7402581 | 10 | Arthropoda | Insecta | Diptera | + | + | |
| TS3.Lobo_1162454 | 2 | Arthropoda | Insecta | Diptera | Chironomus aprilinus | + | + |
| HF4.Lobo_5006 | 1 | Arthropoda | Insecta | Diptera | Cryptochironomus supplicans | + | - |
| TF5.Leray_2910679 | 6 | Arthropoda | Insecta | Diptera | Procladius sp. | + | + |
| HF9.Lobo_7599310 | 3 | Arthropoda | Insecta | Diptera | Psectrocladius yunoquartus | + | + |
| HE5.Lobo_479906 | 152 | Arthropoda | Insecta | Diptera | Tanytarsus usmaensis | + | + |
| HE2.Lobo_2023271 | 21589 | Arthropoda | Malacostraca | Amphipoda | + | + | |
| HF1.Leray_2493444 | 3911 | Arthropoda | Malacostraca | Amphipoda | + | - | |
| HE8.Lobo_860608 | 1 | Arthropoda | Malacostraca | Amphipoda | + | - | |
| HE3.Lobo_4900763 | 1 | Arthropoda | Malacostraca | Amphipoda | Ampelisca brevicornis | + | - |
| HF4.Leray_6193380 | 66039 | Arthropoda | Malacostraca | Amphipoda | Atylus vedlomensis | + | + |
| HE8.Leray_7216397 | 1 | Arthropoda | Malacostraca | Amphipoda | Corophium volutator | + | - |
| HE6.Lobo_7849183 | 1 | Arthropoda | Malacostraca | Amphipoda | Leptocheirus hirsutimanus | + | - |
| HE1.Lobo_914374 | 14588 | Arthropoda | Malacostraca | Amphipoda | Monocorophium insidiosum | + | + |
| TF1.Leray_2445583 | 56 | Arthropoda | Malacostraca | Amphipoda | Monoculodes packardi | - | + |
| TF6.Leray_5321299 | 11588 | Arthropoda | Malacostraca | Cumacea | + | + | |
| HF9.Leray_4291607 | 1372 | Arthropoda | Malacostraca | Decapoda | Athanas nitescens | + | - |
| HF8.Leray_5586003 | 2864 | Arthropoda | Malacostraca | Decapoda | Eualus cranchii | + | + |
| HF8.Leray_5612792 | 37 | Arthropoda | Malacostraca | Decapoda | Eualus cranchii | + | - |
| HE1.Lobo_952576 | 3739 | Arthropoda | Malacostraca | Decapoda | Liocarcinus navigator | + | - |
| TF5.Lobo_6459477 | 1279 | Arthropoda | Malacostraca | Decapoda | Philocheras bispinosus bispinosus | + | + |
| HE4.Lobo_4138563 | 42 | Arthropoda | Malacostraca | Decapoda | Pisidia longicornis | + | + |
| HE8.Leray_7306131 | 2 | Arthropoda | Malacostraca | Decapoda | Processa modica | + | - |
| TS3.Lobo_1213146 | 17 | Arthropoda | Malacostraca | Isopoda | Asellus aquaticus | + | + |
| TF5.Leray_2897128 | 3 | Arthropoda | Maxillopoda | Calanoida | Acartia bifilosa | - | + |
| HF3.Leray_7129076 | 22 | Arthropoda | Maxillopoda | Calanoida | Acartia clausi | + | + |
| TF6.Leray_5332240 | 7399 | Arthropoda | Maxillopoda | Calanoida | Acartia tonsa | + | + |
| HF7.Leray_1683272 | 927 | Arthropoda | Maxillopoda | Calanoida | Acartia tonsa | + | + |
| HE2.Lobo_2010882 | 1 | Arthropoda | Maxillopoda | Calanoida | Anomalocera patersoni | + | - |
| TS2.Leray_4478240 | 2 | Arthropoda | Maxillopoda | Calanoida | Calanus euxinus | - | + |
| HF7.Lobo_5810493 | 41 | Arthropoda | Maxillopoda | Calanoida | Centropages hamatus | + | + |
| HF8.Lobo_5106754 | 82 | Arthropoda | Maxillopoda | Calanoida | Centropages typicus | + | + |
| HE8.Leray_7251655 | 1 | Arthropoda | Maxillopoda | Calanoida | Eurytemora affinis | + | - |
| HE7.Leray_3803390 | 5325 | Arthropoda | Maxillopoda | Calanoida | Paracalanus parvus | + | + |
| HF9.Leray_4411242 | 1 | Arthropoda | Maxillopoda | Calanoida | Pseudocalanus elongatus | + | - |
| TS4.Leray_7515925 | 2 | Arthropoda | Maxillopoda | Calanoida | Pseudocalanus elongatus | - | + |
| TS3.Lobo_1208165 | 1 | Arthropoda | Maxillopoda | Calanoida | Scolecithricella minor | - | + |
| TF5.Lobo_6373065 | 809 | Arthropoda | Maxillopoda | Calanoida | Temora longicornis | + | + |
| TF1.Leray_2453024 | 1 | Arthropoda | Maxillopoda | Calanoida | Temora longicornis | - | + |
| HF4.Leray_6242499 | 45 | Arthropoda | Maxillopoda | Cyclopoida | + | - | |
| HF4.Leray_6206299 | 2 | Arthropoda | Maxillopoda | Harpacticoida | + | - | |
| HE8.Lobo_823478 | 108 | Arthropoda | Maxillopoda | Harpacticoida | Harpacticoida sp. | + | - |
| TS3.Lobo_1208905 | 116 | Arthropoda | Maxillopoda | Harpacticoida | Harpacticus flexus | + | + |
| HE1.Lobo_995710 | 1 | Arthropoda | Maxillopoda | Harpacticoida | Tachidius discipes | + | - |
| HF4.Leray_6092514 | 1 | Arthropoda | Maxillopoda | Poecilostomatoida | + | - | |
| HF9.Leray_4391714 | 11307 | Arthropoda | Maxillopoda | Sessilia | Balanus balanus | + | + |
| HF4.Leray_6295260 | 1079 | Arthropoda | Maxillopoda | Sessilia | Balanus balanus | + | + |
| HF7.Leray_1785147 | 2 | Arthropoda | Maxillopoda | Sessilia | Verruca stroemia | + | - |
| HE1.Leray_1117391 | 1 | Arthropoda | Pycnogonida | Pantopoda | Endeis spinosa | + | - |
| HE9.Lobo_2173983 | 63 | Bryozoa | Gymnolaemata | Cheilostomatida | Escharella immersa | + | - |
| HF7.Leray_1838377 | 98 | Bryozoa | Gymnolaemata | Cheilostomatida | Membranipora membranacea | + | - |
| HE3.Lobo_4881810 | 541 | Bryozoa | Gymnolaemata | Cheilostomatida | Scrupocellaria scruposa | + | - |
| HF6.Lobo_2617384 | 2 | Bryozoa | Gymnolaemata | Ctenostomata | Amathia gracilis | + | - |
| HF5.Lobo_3158598 | 5 | Bryozoa | Stenolaemata | Cyclostomatida | Crisia eburnea | + | - |
| HE6.Leray_2983148 | 31 | Chaetognatha | Sagittoidea | Aphragmophora | + | - | |
| TS1.Leray_646185 | 73 | Chordata | Actinopterygii | Gasterosteiformes | Gasterosteus aculeatus | + | + |
| HF4.Lobo_208606 | 1 | Chordata | Actinopterygii | Perciformes | Ammodytes marinus | + | - |
| HF1.Leray_2487062 | 288 | Chordata | Actinopterygii | Perciformes | Ctenolabrus rupestris | + | - |
| HF3.Lobo_3538759 | 472 | Chordata | Actinopterygii | Perciformes | Gobius niger | + | - |
| TF1.Lobo_2807051 | 486 | Chordata | Actinopterygii | Perciformes | Lesueurigobius friesii | + | + |
| HF9.Lobo_7596943 | 8 | Chordata | Actinopterygii | Perciformes | Mullus surmuletus | + | - |
| HF5.Lobo_3273051 | 43 | Chordata | Actinopterygii | Perciformes | Trachinus draco | + | - |
| HE2.Lobo_1914646 | 81 | Chordata | Actinopterygii | Pleuronectiformes | Limanda limanda | + | - |
| HE8.Lobo_879846 | 265 | Chordata | Actinopterygii | Pleuronectiformes | Solea solea | + | - |
| HE8.Lobo_756051 | 34 | Chordata | Actinopterygii | Salmoniformes | Salmo trutta | + | - |
| HF3.Lobo_3595218 | 14 | Chordata | Ascidiacea | Phlebobranchia | Phallusia ingeria | + | - |
| HE8.Lobo_873511 | 131011 | Chordata | Leptocardii | - | Branchiostoma lanceolatum | + | + |
| TF3.Leray_6588680 | 3869 | Cnidaria | Anthozoa | Pennatulacea | Funiculina sp. | + | + |
| TF6.Lobo_5251371 | 1 | Cnidaria | Hydrozoa | Anthoathecata | Corymorpha nutans | - | + |
| HE9.Lobo_2164485 | 2 | Cnidaria | Hydrozoa | Anthoathecata | Lizzia blondina | + | - |
| TF6.Leray_5512978 | 1481 | Cnidaria | Hydrozoa | Leptothecata | Eutima gracilis | + | + |
| HF5.Lobo_3253786 | 232 | Cnidaria | Scyphozoa | Semaeostomeae | Aurelia aurita | + | + |
| HE3.Leray_361248 | 14 | Cnidaria | Scyphozoa | Semaeostomeae | Cyanea capillata | + | + |
| HE2.Leray_6553538 | 1 | Cnidaria | Staurozoa | Stauromedusae | + | - | |
| HE2.Leray_6571642 | 184 | Cnidaria | Staurozoa | Stauromedusae | Craterolophus convolvulus | + | - |
| HE7.Leray_3802459 | 570 | Echinodermata | Asteroidea | Forcipulatida | Asterias rubens | + | - |
| HE3.Leray_388102 | 85 | Echinodermata | Asteroidea | Forcipulatida | Marthasterias glacialis | + | - |
| HF4.Leray_6293728 | 71 | Echinodermata | Echinoidea | Clypeasteroida | Echinocyamus pusillus | + | + |
| HE8.Leray_7326980 | 315 | Echinodermata | Echinoidea | Echinoida | Psammechinus miliaris | + | - |
| HE6.Lobo_7886165 | 1 | Echinodermata | Echinoidea | Spatangoida | + | - | |
| TF3.Leray_6591339 | 2079 | Echinodermata | Echinoidea | Spatangoida | Brissopsis lyrifera | + | + |
| HF7.Leray_1843674 | 94 | Echinodermata | Echinoidea | Spatangoida | Echinocardium cordatum | + | - |
| TS5.Lobo_6025603 | 11 | Echinodermata | Holothuroidea | Dendrochirotida | Thyone fusus | + | + |
| TS3.Leray_6733304 | 1027065 | Echinodermata | Ophiuroidea | Ophiurida | + | + | |
| TS1.Leray_663710 | 3 | Echinodermata | Ophiuroidea | Ophiurida | Acrocnida brachiata | - | + |
| TF1.Lobo_2726978 | 298 | Echinodermata | Ophiuroidea | Ophiurida | Ophiothrix fragilis | - | + |
| TF1.Leray_2426830 | 16603 | Echinodermata | Ophiuroidea | Ophiurida | Ophiura albida | + | + |
| TF5.Leray_2879711 | 1 | Echinodermata | Ophiuroidea | Ophiurida | Ophiura sarsii | - | + |
| HF3.Leray_7012508 | 44 | Gastrotricha | _ | Macrodasyida | Macrodasys sp. | + | - |
| HE1.Lobo_948618 | 14 | Gnathostomulida | Bursovaginoidea | Gnathostomula armata | + | - | |
| TS2.Leray_4506244 | 1 | Mollusca | Bivalvia | Lucinoida | Thyasira equalis | - | + |
| HF3.Leray_7058438 | 371 | Mollusca | Bivalvia | Myoida | Corbula gibba | + | + |
| HE1.Lobo_894587 | 22 | Mollusca | Bivalvia | Mytiloida | Mytilus edulis | + | - |
| TS1.Lobo_4571224 | 4 | Mollusca | Bivalvia | Nuculida | Nucula nucleus | - | + |
| TS3.Leray_6727248 | 56213 | Mollusca | Bivalvia | Veneroida | Abra nitida | + | + |
| HE4.Lobo_4121128 | 25 | Mollusca | Bivalvia | Veneroida | Dosinia lupinus | + | + |
| TF5.Leray_2915847 | 1911 | Mollusca | Bivalvia | Veneroida | Kurtiella bidentata | + | + |
| TS6.Leray_5683559 | 2 | Mollusca | Bivalvia | Veneroida | Lucinoma borealis | - | + |
| HF1.Leray_2592679 | 33 | Mollusca | Bivalvia | Veneroida | Spisula subtruncata | + | - |
| HE7.Leray_3779267 | 14392 | Mollusca | Bivalvia | Veneroida | Tellimya ferruginosa | + | + |
| HF5.Lobo_3246886 | 1 | Mollusca | Cephalopoda | Sepiida | Sepietta neglecta | + | - |
| TS1.Lobo_4750257 | 2 | Mollusca | Gastropoda | Cephalaspidea | - | + | |
| TS1.Lobo_4792606 | 2 | Mollusca | Gastropoda | Cephalaspidea | - | + | |
| HF8.Lobo_5143779 | 2 | Mollusca | Gastropoda | Littorinimorpha | Euspira nitida | + | - |
| HE3.Lobo_4838288 | 34 | Mollusca | Gastropoda | Neogastropoda | Mangelia attenuata | + | + |
| HF6.Lobo_2622544 | 37 | Mollusca | Gastropoda | Neogastropoda | Nassarius nitidus | + | - |
| HE2.Lobo_1993552 | 50 | Mollusca | Gastropoda | Nudibranchia | + | - | |
| HE6.Leray_2935130 | 2 | Mollusca | Gastropoda | Nudibranchia | + | - | |
| HF1.Leray_2520121 | 559 | Mollusca | Gastropoda | Nudibranchia | Favorinus branchialis | + | - |
| HE2.Lobo_1978270 | 5 | Mollusca | Gastropoda | Nudibranchia | Onchidoris muricata | + | - |
| HE2.Lobo_1939813 | 155 | Mollusca | Gastropoda | Nudibranchia | Polycera quadrilineata | + | - |
| HE2.Lobo_1938412 | 10 | Mollusca | Gastropoda | Nudibranchia | Polycera quadrilineata | + | - |
| HF5.Leray_3991765 | 847 | Mollusca | Gastropoda | Pulmonata | Microhedyle glandulifera | + | - |
| HF4.Leray_6295954 | 2965 | Mollusca | Gastropoda | Sacoglossa | Elysia viridis | + | + |
| HF5.Lobo_3167773 | 166 | Mollusca | Gastropoda | Sorbeoconcha | Onoba semicostata | + | - |
| HE4.Lobo_4138137 | 2 | Mollusca | Gastropoda | Sorbeoconcha | Pusillina inconspicua | + | - |
| TS1.Lobo_4644275 | 2 | Nemertea | Anopla | _ | Cerebratulus sp. | + | + |
| HE4.Lobo_4203493 | 3 | Nemertea | Palaeonemertea | _ | Carinina ochracea | + | - |
| TF1.Lobo_2662495 | 1 | Nemertea | Palaeonemertea | _ | Hubrechtella dubia | - | + |
| HF7.Lobo_5876008 | 353 | Phoronida | _ | _ | Phoronis muelleri | + | - |
| HE8.Lobo_843910 | 13 | Porifera | Demospongiae | Chondrillida | Halisarca dujardini | + | - |
| HE4.Leray_3148053 | 1664 | Porifera | Demospongiae | Suberitida | Halichondria panicea | + | + |
| TS5.Leray_1547671 | 2628 | Priapulida | Priapulimorpha | Priapulimorphida | Priapulus caudatus | + | + |
| HF5.Leray_3885266 | 5 | Rotifera | Eurotatoria | Flosculariaceae | Testudinella clypeata | + | - |
| HE3.Leray_357208 | 2 | Rotifera | Monogononta | Ploima | + | - | |
| HF8.Lobo_5184437 | 1 | Sipuncula | Sipunculidea | Golfingiida | Golfingia vulgaris | + | - |
| TS1.Lobo_4586276 | 14 | Xenacoelomorpha | _ | Acoela | Archaphanostoma sp. | - | + |
| TS3.Lobo_1178177 | 4 | Xenacoelomorpha | _ | Acoela | Childia macroposthium | - | + |
| HF9.Lobo_7719366 | 2 | Xenacoelomorpha | _ | Acoela | Haplogonaria viridis | + | - |
| HF9.Lobo_7734506 | 1 | Xenacoelomorpha | _ | Acoela | Notocelis Gullmarnensis | + | - |
| 18Sa | |||||||
| OTU ID | Nb of reads | Phylum | Class | Order | Species | HI | GF |
| TF5.SSU_460284 | 121639 | Annelida | _ | _ | + | + | |
| TS3.SSU_470635 | 59 | Annelida | _ | _ | - | + | |
| HF9.SSU_7624 | 12 | Annelida | Clitellata | Enchytraeida | Grania sp. | + | - |
| TF5.SSU_453927 | 2687 | Annelida | Clitellata | Haplotaxida | Tubificoides insularis | + | + |
| HF3.SSU_985477 | 1090 | Annelida | Polychaeta | _ | Aricia sp. | + | + |
| HF6.SSU_322303 | 10 | Annelida | Polychaeta | _ | Protodriloides chaetifer | + | - |
| HF4.SSU_622170 | 1 | Annelida | Polychaeta | _ | Scalibregma inflatum | + | - |
| HF9.SSU_25735 | 3753 | Annelida | Polychaeta | _ | Trilobodrilus heideri | + | - |
| TS3.SSU_480632 | 189 | Annelida | Polychaeta | Phyllodocida | Aphrodita sp. | - | + |
| HE6.SSU_371492 | 49226 | Annelida | Polychaeta | Phyllodocida | Brania sp. | + | + |
| HE4.SSU_913344 | 37252 | Annelida | Polychaeta | Phyllodocida | Glycera sp. | + | + |
| HF5.SSU_997904 | 64 | Annelida | Polychaeta | Phyllodocida | Glycinde armigera | + | + |
| TS5.SSU_870099 | 69 | Annelida | Polychaeta | Phyllodocida | Goniada maculata | - | + |
| TF6.SSU_42415 | 2 | Annelida | Polychaeta | Phyllodocida | Harmothoe imbricata | - | + |
| HE6.SSU_350003 | 5 | Annelida | Polychaeta | Phyllodocida | Myrianida sp. | + | - |
| HF6.SSU_324605 | 2 | Annelida | Polychaeta | Phyllodocida | Nereis pelagica | + | - |
| HE7.SSU_239005 | 67220 | Annelida | Polychaeta | Phyllodocida | Pisione remota | + | + |
| HE2.SSU_637269 | 49 | Annelida | Polychaeta | Phyllodocida | Platynereis dumerilii | + | - |
| HE8.SSU_832291 | 1 | Annelida | Polychaeta | Phyllodocida | Progoniada regularis | + | - |
| HE8.SSU_834197 | 1 | Annelida | Polychaeta | Sabellida | Fabriciola liguronis | + | - |
| HF2.SSU_202737 | 4 | Annelida | Polychaeta | Sabellida | Laeospira corallinae | + | - |
| HE2.SSU_640060 | 3 | Annelida | Polychaeta | Sabellida | Myriochele sp. | + | - |
| TS5.SSU_869292 | 123 | Annelida | Polychaeta | Spionida | Apistobranchus sp. | - | + |
| TS3.SSU_517096 | 1407 | Annelida | Polychaeta | Spionida | Laonice sp. | - | + |
| HE3.SSU_123438 | 1952 | Annelida | Polychaeta | Spionida | Spio sp. | + | + |
| TS5.SSU_882766 | 60 | Annelida | Polychaeta | Terebellida | Diplocirrus glaucus | - | + |
| HF2.SSU_193854 | 1 | Annelida | Polychaeta | Terebellida | Flabelligera sp. | + | - |
| TF6.SSU_63146 | 669 | Annelida | Polychaeta | Terebellida | Pectinaria sp. | - | + |
| TS5.SSU_883475 | 4155 | Annelida | Polychaeta | Terebellida | Terebellides stroemii | - | + |
| TF4.SSU_139713 | 193 | Arthropoda | Branchiopoda | _ | - | + | |
| HE5.SSU_184679 | 149 | Arthropoda | Malacostraca | _ | + | - | |
| HE8.SSU_832214 | 1 | Arthropoda | Malacostraca | Decapoda | Nikoides sp. | + | - |
| HF5.SSU_994971 | 7 | Arthropoda | Malacostraca | Decapoda | Praebebalia longidactyla | + | - |
| TF6.SSU_56595 | 65992 | Arthropoda | Maxillopoda | _ | + | + | |
| HF9.SSU_15855 | 31800 | Arthropoda | Maxillopoda | _ | + | + | |
| HF2.SSU_208480 | 21241 | Arthropoda | Maxillopoda | _ | + | + | |
| TS2.SSU_812824 | 433 | Arthropoda | Maxillopoda | _ | + | + | |
| TF3.SSU_955499 | 185 | Arthropoda | Maxillopoda | _ | + | + | |
| TF5.SSU_470101 | 360 | Arthropoda | Maxillopoda | Harpacticoida | Typhlamphiascus typhlops | - | + |
| HE1.SSU_864375 | 1160 | Arthropoda | Ostracoda | Podocopida | Hemicytherura kajiyamai | + | + |
| HE7.SSU_253407 | 2584 | Arthropoda | Ostracoda | Podocopida | Loxocorniculum mutsuense | + | + |
| HE5.SSU_181011 | 1 | Arthropoda | Pycnogonida | Pantopoda | Anoplodactylus californicus | + | - |
| HE2.SSU_646490 | 123 | Arthropoda | Pycnogonida | Pantopoda | Callipallene sp. | + | - |
| HE2.SSU_638224 | 23 | Bryozoa | _ | _ | + | - | |
| HE6.SSU_373369 | 2 | Bryozoa | Stenolaemata | Cyclostomatida | Plagioecia patina | + | - |
| HE1.SSU_850917 | 4 | Bryozoa | Stenolaemata | Cyclostomatida | Tubulipora lobifera | + | - |
| TF5.SSU_412099 | 18 | Cephalorhyncha | Kinorhyncha | Homalorhagida | Pycnophyes kielensis | - | + |
| HE7.SSU_239963 | 45 | Chordata | Actinopteri | Perciformes | Hypseleotris sp. | + | + |
| HE3.SSU_123107 | 4 | Chordata | Ascidiacea | _ | + | - | |
| HF9.SSU_12142 | 727 | Chordata | Ascidiacea | Phlebobranchia | Ascidiella sp. | + | + |
| HF4.SSU_611685 | 114 | Chordata | Ascidiacea | Phlebobranchia | Corella inflata | + | + |
| HE2.SSU_639404 | 209 | Chordata | Ascidiacea | Stolidobranchia | Molgula sp. | + | - |
| HE9.SSU_314754 | 616 | Chordata | Ascidiacea | Stolidobranchia | Styela plicata | + | - |
| HE8.SSU_834024 | 11058 | Chordata | Leptocardii | _ | Branchiostoma sp. | + | - |
| TF1.SSU_674740 | 2212 | Cnidaria | Anthozoa | Actiniaria | Nematostella vectensis | + | + |
| TS3.SSU_472524 | 2741 | Cnidaria | Hydrozoa | _ | + | + | |
| TS3.SSU_518760 | 7860 | Cnidaria | Hydrozoa | Anthoathecata | Euphysa sp. | + | + |
| HE2.SSU_639670 | 1 | Cnidaria | Hydrozoa | Leptothecatha | Abietinaria filicula | + | - |
| TF4.SSU_152912 | 61418 | Echinodermata | _ | _ | + | + | |
| HE5.SSU_186025 | 8038 | Echinodermata | _ | _ | + | + | |
| TF4.SSU_155631 | 5491 | Echinodermata | _ | _ | + | + | |
| TS5.SSU_881395 | 25 | Echinodermata | _ | _ | - | + | |
| HE4.SSU_914821 | 1 | Echinodermata | Holothuroidea | Apodida | Leptosynapta sp. | + | - |
| HF9.SSU_2577 | 1006 | Gastrotricha | _ | Chaetonotida | Chaetonotus sp. | + | + |
| HE7.SSU_244283 | 249 | Gastrotricha | _ | Macrodasyida | Diplodasys meloriae | + | - |
| HF5.SSU_996540 | 161 | Gastrotricha | _ | Macrodasyida | Lepidodasys sp. | + | - |
| HF5.SSU_995416 | 636 | Gastrotricha | _ | Macrodasyida | Macrodasys sp. | + | - |
| HF2.SSU_192734 | 479 | Gastrotricha | _ | Macrodasyida | Macrodasys sp. | + | - |
| HF7.SSU_385728 | 6934 | Gastrotricha | _ | Macrodasyida | Mesodasys sp. | + | + |
| HE7.SSU_242889 | 3013 | Gastrotricha | _ | Macrodasyida | Tetranchyroderma thysanophorum | + | - |
| HF1.SSU_770513 | 339 | Gastrotricha | _ | Macrodasyida | Thaumastoderma ramuliferum | + | - |
| HF1.SSU_760431 | 5 | Gastrotricha | _ | Macrodasyida | Urodasys sp. | + | - |
| TF6.SSU_44832 | 3816 | Mollusca | Bivalvia | _ | + | + | |
| HF2.SSU_208561 | 14 | Mollusca | Bivalvia | Anomalodesmata | + | + | |
| HF8.SSU_788507 | 1 | Mollusca | Bivalvia | Limoida | Limaria hians | + | - |
| TF3.SSU_924397 | 11725 | Mollusca | Bivalvia | Veneroida | Abra sp. | + | + |
| HE9.SSU_317977 | 1982 | Mollusca | Bivalvia | Verenoida | Arctica islandica | + | + |
| TF4.SSU_132537 | 1581 | Mollusca | Gastropoda | Neogastropoda | Nassarius festivus | + | + |
| HF1.SSU_779114 | 65 | Nematoda | Chromadorea | Araeolaimida | Odontophora sp. | + | + |
| TF6.SSU_48167 | 2940 | Nematoda | Chromadorea | Araeolaimida | Sabatieria sp. | + | + |
| TF1.SSU_710679 | 639 | Nematoda | Chromadorea | Chromadorida | + | + | |
| HF2.SSU_192072 | 2 | Nematoda | Chromadorea | Chromadorida | Chromadora nudicapitata | + | - |
| HF1.SSU_759758 | 4 | Nematoda | Chromadorea | Plectida | + | - | |
| HF9.SSU_20251 | 636 | Nematoda | Desmodorida | Microlaimidae | + | + | |
| HE3.SSU_124287 | 13 | Nematoda | Enoplea | Enoplida | Enoploides sp. | + | - |
| HE3.SSU_110275 | 8 | Nematoda | Enoplea | Enoplida | Enoplus sp. | + | - |
| HE5.SSU_188855 | 27 | Nematoda | Enoplea | Enoplida | Symplocostoma sp. | + | + |
| TS6.SSU_587229 | 493 | Nematoda | Enoplea | Enoplida | Viscosia viscosa | + | + |
| TF3.SSU_938615 | 642 | Nemertea | _ | _ | + | + | |
| TF6.SSU_49192 | 265 | Nemertea | Anopla | _ | Cerebratulus marginatus | + | + |
| HE4.SSU_908113 | 877 | Nemertea | Anopla | _ | Lineus bilineatus | + | + |
| HF9.SSU_3582 | 6 | Nemertea | Paleonemertea | _ | Callinera grandis | + | - |
| HE3.SSU_121696 | 12053 | Nemertea | Paleonemertea | _ | Cephalothrix filiformis | + | + |
| TF5.SSU_434928 | 1760 | Nemertea | Paleonemertea | _ | Hubrechtella dubia | + | + |
| TS2.SSU_818002 | 1 | Platyhelminthes | Rhabditophora | Cestoda | - | + | |
| HE9.SSU_303121 | 1939 | Platyhelminthes | Rhabditophora | Haplopharyngida | Haplopharynx rostratus | + | - |
| HF1.SSU_773830 | 1 | Platyhelminthes | Rhabditophora | Prolecithophora | Allostoma neostiliferum | + | - |
| HE2.SSU_650311 | 8 | Platyhelminthes | Rhabditophora | Prolecithophora | Cylindrostoma sp. | + | - |
| HE5.SSU_177399 | 4 | Platyhelminthes | Rhabditophora | Prolecithophora | Euxinia baltica | + | - |
| HF9.SSU_23023 | 8367 | Platyhelminthes | Rhabditophora | Prolecithophora | Plagiostomum cinctum | + | + |
| TS2.SSU_822141 | 938 | Platyhelminthes | Rhabditophora | Prolecithophora | Plagiostomum cuticulata | - | + |
| TF6.SSU_52738 | 214 | Platyhelminthes | Rhabditophora | Prolecithophora | Plagiostomum striatum | - | + |
| TF5.SSU_433159 | 2 | Platyhelminthes | Rhabditophora | Prolecithophora | Ulianinia mollissima | - | + |
| HF9.SSU_24513 | 59 | Platyhelminthes | Rhabditophora | Proseriata | Monocelis lineata | + | + |
| HF2.SSU_201740 | 2 | Platyhelminthes | Rhabditophora | Rhabdocoela | Phonorhynchus helgolandicus | + | - |
| TS6.SSU_592673 | 245 | Platyhelminthes | Rhabditophora | Rhabdocoela | Proxenetes sp. | + | + |
| HF4.SSU_616041 | 771 | Platyhelminthes | Rhabditophora | Seriata | + | - | |
| HE3.SSU_117223 | 181 | Porifera | Calcarea | _ | + | + | |
| HE7.SSU_223989 | 12 | Porifera | Demospongiae | Chondrillida | Halisarca dujardini | + | - |
| HF9.SSU_26977 | 8 | Porifera | Demospongiae | Clionaida | Spheciospongia vesparium | + | - |
| HE6.SSU_383060 | 3 | Sipuncula | Sipunculidea | Golfingiida | Phascolopsis gouldii | + | - |
| HE6.SSU_348954 | 2 | Tardigrada | Eutardigrada | Parachela | Halobiotus crispae | + | - |
| TF3.SSU_927927 | 2 | Xenacoelomorpha | _ | _ | - | + | |
| HE3.SSU_116025 | 28 | Xenacoelomorpha | _ | Acoela | Archaphanostoma sp. | + | + |
| HF9.SSU_26335 | 1 | Xenacoelomorpha | _ | Acoela | Archaphanostoma sp. | + | - |
| TS2.SSU_815721 | 2 | Xenacoelomorpha | _ | Acoela | Childia sp. | - | + |
| TS2.SSU_815970 | 1 | Xenacoelomorpha | _ | Acoela | Childia sp. | - | + |
| HF2.SSU_190395 | 2386 | Xenacoelomorpha | _ | Acoela | Eumecynostomum sp. | + | - |
| HF1.SSU_758202 | 74 | Xenacoelomorpha | _ | Acoela | Haplogonaria sp. | + | + |
| HF9.SSU_13290 | 5 | Xenacoelomorpha | _ | Nemertodermatida | Flagellophora apelti | + | - |
| TS6.SSU_601153 | 28 | Xenacoelomorpha | _ | Nemertodermatida | Nemertoderma westbladi | - | + |
Invasive and alien species detected in the samples
Five alien species were detected in in the sample, of which two are considered invasive (in bold; Table
Invasive species (in bold) and species on alert lists (not bold) found in the samples. X indicates where the species were found.
|
Species |
Phylum |
COI |
18S |
||
|
Hållö island |
Gullmarn Fjord |
Hållö island |
Gullmarn Fjord |
||
|
Acartia tonsa |
Arthropoda |
x |
x |
|
|
|
Alexandrium ostenfeldii |
Dinoflagellata |
|
|
x |
x |
|
Bonnemaisonia hamifera |
Rhodophyta |
x |
x |
x |
|
|
Penilia avirostris |
Arthropoda |
x |
x |
|
|
|
Thalassiosira punctigera |
Bacillariophyta |
x |
|
|
|
Comparison of metabarcoding versus morphology-based identification of Xenacoelomorpha
Comparison of morphology-based assessment of Xenacoelomorpha diversity with metabarcoding using taxonomic assignments to the phylum level (with 80% similarity threshold; Suppl. materials
Taxonomic composition and relative abundance (% of the total number of specimens) of Xenacoelomorpha species in Gullmarn Fjord and Hållö sites.
|
|
|
Gullmarn Fjord |
Hållö |
||
|
|
Taxon |
Siphoning |
Flotation with fresh water |
Flotation with MgCl2 solution |
Flotation with fresh water |
|
|
Acoela |
|
|
|
|
|
1 |
Haploposthia rubropunctata |
1.03 |
0 |
0 |
0 |
|
2 |
Childia brachyposthium |
3.78 |
0 |
0 |
0 |
|
3 |
Childia submaculatum |
1.03 |
0 |
0 |
0 |
|
4 |
Childia trianguliferum |
2.06 |
0 |
0 |
0 |
|
5 |
Childia crassum |
3.44 |
0 |
0 |
0 |
|
6 |
Childia sp. |
25.09 |
0 |
0 |
0 |
|
7 |
Mecynostomum tenuissimum |
43.99 |
0 |
0 |
0 |
|
8 |
Mecynostomum auritum |
0.34 |
0 |
0 |
0 |
|
9 |
cf. Eumecynostomum altitudi |
4.81 |
0 |
0 |
0 |
|
10 |
Philactinoposthia sp. |
0.34 |
0 |
0 |
0 |
|
11 |
Acoela sp. |
2.06 |
100 |
88.71 |
0 |
|
12 |
Faerlea glomerata |
3.09 |
0 |
|
|
|
13 |
Archaphanostoma sp. |
0.34 |
0 |
0.81 |
0 |
|
14 |
Postmecynostomum glandulosum |
0 |
0 |
2.42 |
0 |
|
15 |
Paramecynostomum sp. |
0 |
0 |
0.81 |
0 |
|
16 |
Eumecynostomum macrobursalium |
0 |
0 |
0.81 |
0 |
|
17 |
Isodiametra sp. |
0 |
0 |
0.81 |
0 |
|
18 |
Haplogonaria viridis/Archocelis macrorhabditis |
0 |
0 |
5.65 |
0 |
|
|
Nemertodermatida |
|
|
|
|
|
19 |
Nemertoderma westbladi |
8.25 |
0 |
0 |
0 |
|
20 |
Flagellophora apelti |
0.34 |
0 |
0 |
0 |
Total number of Xenacoelomorpha taxa or OTUs distinguished based on morphology (Table
|
Site / extraction method |
morphology-based |
18S |
COI (Lobo) |
COI (Leray) |
|
Hållo, flotation with MgCl2 |
7 |
11 |
8 |
6 |
|
Hållö, flotation with fresh water |
0 |
15 |
11 |
6 |
|
Hållö, total |
7 |
16 |
12 |
7 |
|
Gullmarn Fjord, siphoning |
15 |
11 |
9 |
4 |
|
Gullmarn Fjord, flotation with fresh water |
1 |
13 |
2 |
0 |
|
Gullmarn Fjord, total |
15 |
19 |
10 |
4 |
Comparison of metabarcoding versus morphology-based identification of Nematoda
Both study sites are characterized by rich and diverse nematode fauna. The Hållö site had a total of 107 species of nematodes, belonging to 86 genera (
The final list of nematode OTUs includes 139 18S sequences. Only two 18S OTUs were positively identified using QIIME to species level using 97% similarity threshold: Viscosia viscosa (TS6.SSU58722) and Chromadora nudicapitata (HF2.SSU192072), six more were assigned to reference sequences identified to genus level only (Suppl. material
When comparing the results of morphology-based assessment of nematode diversity with metabarcoding using taxonomic assignments to the phylum level in this particular study (with 80% similarity threshold; Suppl. materials
Total number of nematode taxa or OTUs distinguished based on morphology (after
|
Site / extraction method |
morphology-based |
18S |
COI (Lobo) |
COI (Leray) |
|
Hållo, flotation with MgCl2 |
88 |
71 |
12 |
11 |
|
Hållö, flotation with fresh water |
101 |
78 |
14 |
14 |
|
Hållö, total |
107 |
95 |
16 |
17 |
|
Gullmarn Fjord, siphoning |
81 |
47 |
8 |
4 |
|
Gullmarn Fjord, flotation with fresh water |
102 |
67 |
4 |
2 |
|
Gullmarn Fjord, total |
113 |
78 |
9 |
4 |
Acknowledgements
We would like to thank the Genomics Core facility platform at the Sahlgrenska Academy, University of Gothenburg. The SweBoL (Swedish Barcode of Life) network and Christer Erséus are thanked for sharing barcode databases of Swedish invertebrates. We would also like to thank Nicolas Girard for help with scripting. This work was in part supported by the project "Systematics of Swedish free-living nematodes of the orders Desmodorida and Araeolaimida" (Swedish Taxonomy Initiative, ArtDatabanken, Sweden) awarded to OH, and by the Swedish Research Council project (2012-3446) 'Biodiversity genomics: Species identification pipelines for analyzing marine invertebrate larval stages, community structure, and trophic interactions’ awarded to SJB.
References
- Hidden diversity of Acoelomorpha revealed through metabarcoding.Biology Letters12(9):20160674. https://doi.org/10.1098/rsbl.2016.0674
- The Genus Alexandrium Halim (Dinoflagellata).Sherkin Island Marine Station, IrelandISBN: 1 870492 61 7.
- Dramatic Shifts in Benthic Microbial Eukaryote Communities following the Deepwater Horizon Oil Spill.Plos One7(6):e38550. [InEnglish]. https://doi.org/10.1371/journal.pone.0038550
- Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments.Molecular Ecology21(5):1048‑1059. [InEnglish]. https://doi.org/10.1111/j.1365-294X.2011.05297.x
- Environmental DNA for wildlife biology and biodiversity monitoring.Trends in ecology & evolution29(6):358‑67. https://doi.org/10.1016/j.tree.2014.04.003
- Bourlat SJ, Haenel Q, Finnman J, Leray M (2016) Metabarcoding of marine eukaryotes: Illumina Mi-Seq library preparation using fusion primer methods. In: Bourlat SJ (Ed.) Marine Genomics - Methods and protocols.Springer,New York. [ISBN978-1-4939-3772-1].
- Meiofaunal community analysis by high-throughput sequencing: Comparison of extraction, quality filtering, and clustering methods.Marine GenomicsIn Press.
- QIIME allows analysis of high-throughput community sequencing data.Nat Methods7(5):335‑6. [Ineng]. https://doi.org/10.1038/nmeth.f.303
- Metabarcoding Is Powerful yet Still Blind: A Comparative Analysis of Morphological and Molecular Surveys of Seagrass Communities.Plos One10(2):e0117562. [InEnglish]. https://doi.org/10.1371/Journal.Pone.0117562
- Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises.Molecular Ecology19:4‑20. [InEnglish]. https://doi.org/10.1111/J.1365-294x.2009.04473.X
- Redescription ou modifications de quelques techniques utilisées dans l'etude des nematodes phytoparasitaires.Mededelingen Rijksfakulteit Landbouwwetenschappen Gent34:351‑369.
- Eukaryotic plankton diversity in the sunlit ocean.Science348(6237). [InEnglish]. https://doi.org/10.1126/Science.1261605
- Search and clustering orders of magnitude faster than BLAST.Bioinformatics26(19):2460‑1. https://doi.org/10.1093/bioinformatics/btq461
- UCHIME improves sensitivity and speed of chimera detection.Bioinformatics27(16):2194‑200. https://doi.org/10.1093/bioinformatics/btr381
- An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform.Microbiome2(1). https://doi.org/10.1186/2049-2618-2-6
- DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates.Mol Mar Biol Biotechnol3(5):294‑9. [Ineng]. URL: http://www.ncbi.nlm.nih.gov/pubmed/7881515
- Metagenetic analysis of patterns of distribution and diversity of marine meiobenthic eukaryotes.Global Ecology and Biogeography23(11):1293‑1302. [InEnglish]. https://doi.org/10.1111/geb.12223
- Second-generation environmental sequencing unmasks marine metazoan biodiversity.Nature Communications1[InEnglish]. https://doi.org/10.1038/Ncomms1095
- TaxCollector: Modifying Current 16S rRNA Databases for the Rapid Classification at Six Taxonomic Levels.Diversity2(7):1015‑1025. https://doi.org/10.3390/d2071015
- Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering.Bioinformatics27(5):611‑618. [InEnglish]. https://doi.org/10.1093/Bioinformatics/Btq725
- Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species.Proceedings of the Royal Society of London Series B-Biological Sciences270[InEnglish]. https://doi.org/10.1098/Rsbl.2003.0025
- Introduction to the study of Meiofauna.Smithsoniam Institution Press, Washington D.C., London
- The choice of taxonomy assignment approach has strong impact on the efficiency of identification of anonymous metabarcodes of marine nematodes.Manuscript in preparation.
- NOBANIS – Invasive Alien Species Fact Sheet: Acartia tonsa From: Identification key to marine invasive species in Nordic waters.www.nobanis.org. Accessed on: 2017-3-01.
- Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Bioinformatics28(12):1647‑9. https://doi.org/10.1093/bioinformatics/bts199
- Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform.Nucleic Acids Res40(1). https://doi.org/10.1093/nar/gkr771
- Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems.Isme Journal9(5):1208‑1221. [InEnglish]. https://doi.org/10.1038/ismej.2014.213
- CREST – Classification Resources for Environmental Sequence Tags.PLoS ONE7(11):e49334. https://doi.org/10.1371/journal.pone.0049334
- How will the ‘molecular revolution’ contribute to biological recording?Biological Journal of the Linnean Society115(3):750‑766. https://doi.org/10.1111/bij.12516
- DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity.Proceedings of the National Academy of Sciences of the United States of America112(7):2076‑2081. [InEnglish]. https://doi.org/10.1073/pnas.1424997112
- A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents.Frontiers in Zoology10[InEnglish]. https://doi.org/10.1186/1742-9994-10-34
- Next Generation Sequencing Reveals the Hidden Diversity of Zooplankton Assemblages.Plos One8(11):e81327. [InEnglish]. https://doi.org/10.1371/journal.pone.0081327
- Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans.BMC Ecol13https://doi.org/10.1186/1472-6785-13-34
- The use of meiofauna diversity as an indicator of pollution in harbours.Ices Journal of Marine Science65(8):1428‑1435. [InEnglish]. https://doi.org/10.1093/icesjms/fsn116
- Evaluating high-throughput sequencing as a method for metagenomic analysis of nematode diversity.Mol Ecol Resour9(6):1439‑50. https://doi.org/10.1111/j.1755-0998.2009.02611.x
- Next-generation sequencing technologies for environmental DNA research.Molecular Ecology21(8):1794‑1805. [InEnglish]. https://doi.org/10.1111/J.1365-294x.2012.05538.X
- Vincx M (1996) Meiofauna in marine and freshwater sediments. In: Hall GS (Ed.) Methods for the examination of organismal diversity in soils and sediments.
- EvolView, an online tool for visualizing, annotating and managing phylogenetic trees.Nucleic Acids Res40:569‑72. https://doi.org/10.1093/nar/gks576
Supplementary materials
Sequence similarity search at 97% similarity allowed us to identify some OTUs to species level. 215 COI OTUs and 243 18S OTUs were identified to species from both sites (Hållö island and Gullmarsfjord).
OTU table showing all 18S OTUs, their taxonomic assignment at 80% similarity and number of reads per sample (HE: Hållö Flotation, HF: Hållö Flotation MgCl2, TS: Gullmarn Fjord Siphoning, TF: Gullmarn Fjord Flotation)
OTU table showing all COI OTUs, their taxonomic assignment at 80% similarity and number of reads per sample (HE: Hållö Flotation, HF: Hållö Flotation MgCl2, TS: Gullmarn Fjord Siphoning, TF: Gullmarn Fjord Flotation)