|
Biodiversity Data Journal :
Research Article
|
|
Corresponding author: Antonio S. Ortiz (aortiz@um.es)
Academic editor: Bong-Kyu Byun
Received: 26 Jul 2017 | Accepted: 02 Aug 2017 | Published: 08 Aug 2017
© 2017 Antonio Ortiz, Rosa Rubio, Juan Guerrero, Manuel Garre, Jose Serrano, Paul Hebert, Axel Hausmann
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Ortiz A, Rubio R, Guerrero J, Garre M, Serrano J, Hebert P, Hausmann A (2017) Close congruence between Barcode Index Numbers (bins) and species boundaries in the Erebidae (Lepidoptera: Noctuoidea) of the Iberian Peninsula. Biodiversity Data Journal 5: e19840. https://doi.org/10.3897/BDJ.5.e19840
|
|
Abstract
The DNA barcode reference library for Lepidoptera holds much promise as a tool for taxonomic research and for providing the reliable identifications needed for conservation assessment programs. We gathered sequences for the barcode region of the mitochondrial cytochrome c oxidase subunit I gene from 160 of the 176 nominal species of Erebidae moths (Insecta: Lepidoptera) known from the Iberian Peninsula. These results arise from a research project which constructing a DNA barcode library for the insect species of Spain. New records for 271 specimens (122 species) are coupled with preexisting data for 38 species from the Iberian fauna. Mean interspecific distance was 12.1%, while the mean nearest neighbour divergence was 6.4%. All 160 species possessed diagnostic barcode sequences, but one pair of congeneric taxa (Eublemma rosea and Eublemma rietzi) were assigned to the same BIN. As well, intraspecific sequence divergences higher than 1.5% were detected in four species which likely represent species complexes. This study reinforces the effectiveness of DNA barcoding as a tool for monitoring biodiversity in particular geographical areas and the strong correspondence between sequence clusters delineated by BINs and species recognized through detailed taxonomic analysis.
Keywords
barcode library, CO1, Lepidoptera, DNA barcoding, Spain, Iberian Peninsula, mitochondrial DNA
Introduction
The Mediterranean peninsulas of Iberia, Italy, and the Balkans are important hotspots of biodiversity (
Lepidoptera is one of the most species-rich orders of insects, with some 155,000 described species found in diverse habitats from cooler regions to tropical forests (
Since DNA barcodes were proposed as a tool for species identification (
The Iberian macromoth fauna has been well studied taxonomically and ecologically, reflecting its occupation of a peninsular refuge and a bridge between Europe and Northern Africa. This project represents the first in a series that will assemble a DNA barcode library for all macromoth species from the Iberian Peninsula because its species richness and genetic diversity are the highest in Europe.
The present study has the primary goal of providing access to a comprehensive barcode library for the Erebidae species of the Iberian Peninsula. We additionally test how the molecular delineation of COI (mitochondrial cytochrome c oxidase subunit I) barcode haplotype clusters compares with the morphological species concept are useful tools for assessing biodiversity and indicating the completeness of biotic surveys. Such data releases in the Barcode of Life Datasystem (BOLD) and GenBank help to democratize access to biodiversity information because each barcode record is accompanied by georeferenced data and images of its source specimen (
The specific aims of this study are (a) to present a public data release of DNA barcodes for Iberian Erebidae, (b) to critically analyse intraspecific variation and interspecific distances in the barcode region and how they relate to traditionally recognized species and, (c) to test the correspondence between BINs and traditionally recognized species.
Material and methods
Sampling
Specimens were sampled across Spain and the Canary Islands. Permission to collect Lepidoptera in Spain is required both inside and outside nature reserves in all regions. This study considers all 13 subfamilies of Erebidae known from Iberian Peninsula, representing 176 species (
DNA barcodes were obtained by sampling a dry leg from each of a few vouchers per species, trying to include material from all Iberian faunal regions. In total, tissue samples from 271 Iberian specimens (including one Canarian), representing 122 of the species present in the Iberian Peninsula were submitted for analysis. In addition, existing sequence records were included for 38 of the 54 missing species, adding 87 sequences. These samples derived from Germany (67 sequences; 25 species), Italy (13 seqs; 9 spp.) France (2 seqs; 1 sp.), Cyprus, Ethiopia, Hungary, Macedonia and Russia (each 1 seq.; 1 sp.).
DNA Analysis
PCR amplification and DNA sequencing were performed at the Canadian Centre for DNA Barcoding following standard high-throughput protocols (
Data Analysis
Sequence divergences for the barcode region were quantified using the Kimura 2 Parameter distance model, employing the analytical tools in BOLD (BOLD alignment, pairwise deletion). Genetic distances between species are reported as minimum pairwise distances, while intraspecific variation is reported as mean and maximum pairwise distances.
Each specimen with a sequence longer than 500bp (listed in Suppl. material
Results
Traditional and BIN species delineations
Sequences were recovered friom 271 of the 304 (89.5%) specimens. All sequences were longer than 500 bp, meeting the length requirement for DNA barcode status (
The 160 morphological species were assigned to 163 BINs and could be separated into three categories. Most (96.3%) taxa showed a perfect match between morphological species and BINs (154 species). Four (2.5%) species were each placed into two BINs, while just two species, Eublemma rosea and its allopatric congener Eublemma rietzi, were merged into the same BIN. However, even in this case, shallow barcode divergence (2.02%) allows the discrimination of E. rietzi, described in 2010, from the similar but morphologically separable E. rosea.
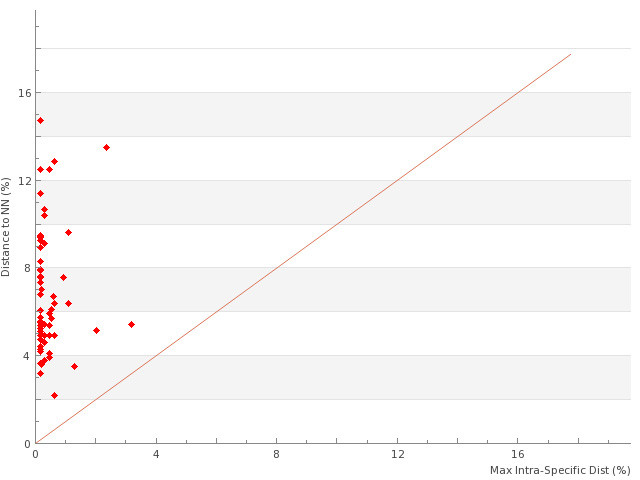
Considering all specimens from all species in the 13 subfamilies, Iberian Erebidae showed a mean interspecific genetic distance of 12.1% (SE <0.01; n=61,901 comparisons of barcodes >500bp). By comparison, congeneric species averaged 6.7% divergence (SE <0.01; n=1,831), while the mean nearest neighbour divergence was 6.4% (n=160). Mean and maximum intraspecific variation were 0.6% and 4.1% respectively, based upon traditionally delimited species including the four assigned to more than one BIN (n=122 species represented by more than one specimen). By comparison, the mean and maximum intra-BIN variation were 0.21% and 3.2% respectively (n=165 BINs represented by more than one specimen; SE=0.01). As a consequence, there was a clear barcode gap for almost all the species (Fig.
Species assigned to multiple BINs
Although most Iberian Erebidae showed very limited intraspecific barcode variation, 4 of the 160 species (see Suppl. material
Among the four species assigned to more than one BIN, 4 of the 8 intraspecific BIN clusters represent cases where both BINs occur in sympatry as defined by cases where the minimum geographic distance between members of the two BINs was less than 100 km. Two cases of BIN splits corresponded to subspecies recognized by traditional taxonomy: Ocnogyna z. zoraida (BOLD:ACE3052) & Ocnogyna z. hemigena (Graslin, 1850) (BOLD:AAY5665), and Arctia v. villica (BOLD:ACP7477) & A. v. angelica (Boisduval, 1829) (BOLD:ABY6789). However, no morphological differences (e.g. wing colour, wing pattern, morphology of genitalia) were evident between members of different BINs in the other two species (Orgyia dubia, Ocnogyna baetica). However, both cases involved geographically isolated lineages with the minimum geographic distance between members of the two BINs being around 125 km in O. dubia and 330 km in O. baetica.
Discussion
Identification accuracy
Following the definition of ‘diagnostic’ barcode clusters including those with monophyletic intraspecific splits (
The success of re-identification by DNA barcoding remains high in Erebidae even when analysis is extended to a European scale. A different pattern with lower number of BIN-species-matches species was reported for European Geometridae (
Different species assigned to the same BIN
Just one pair of species shared a BIN, the recently separated Eublemma rosea and E. rietzi (
A second potential case of BIN sharing involved the Eilema complana/ pseudocomplana complex as its members were allocated to one or two BINs depending on the taxonomic status of certain Iberian populations.
One species assigned to two imore BINs
Because about one fifth (34/160) of the species examined in this study were represented by a single specimen, additional samples, particularly from new geographic areas and habitats will likely reveal new BINs for some Iberian Erebidae. However, the current analysis revealed BIN splits in four species (Rivula sericealis, Eilema sororcula, Coscinia cribraria, Lygephila craccae) represented by singletons show a different haplotype from the rest of the European populations, potentially pointing to taxonomic implications. We detected one species, Ocnogyna baetica, whose Iberian members were assigned to two BINs with divergence greater than 3%, in the same way that Iberian Eublemma parva and Coscinia cribraria singletons present with other conspecific Iberian populations, suggesting these require further study.
Other cases of multiple BINs involved taxa whose discrimination is sometimes uncertain. Ocnogyna z. zoraida and O. z. hemigena as well as Arctia v. villica and A. v. angelica each involve a pair of subspecies of uncertain status. Prior studies have detected specimens with intermediate characters in putative hybrid zones suggesting recent speciation with incomplete lineage sorting and introgression. These lineages occur in sympatry, and usually possess consistent differences in external appearance as noted in the literature or that were apparent from our investigation. These correlations may justify the upgrading of these taxa to a species rank though this decision requires further integrated taxonomic study.
In one BIN-split, Orgyia dubia, the two BINs occurred in allopatry. The presence of two barcode lineages in O. dubia (‘subspecies splendida Rambur, 1842’) with 2.25% divergence between central and southern populations might reflect divergence that arose when populations were isolated in different glacial refugia during the Pleistocene. Moreover, the Iberian subspecies splendida needs to be compared to the nominotypical O. d. dubia Tauscher, 1806 from its type locality in Russia.
The genus Setina is considered taxonomically difficult due to high inter-populational variability in morphology. In our study, Setina flavicans and S. cantabrica showed clear divergence (2.34%) although the nearest neighbour for both taxa is S. irrorella (Linnaeus, 1758) with 1.77% distance. Current taxonomy views both S. cantabrica and S. flavicans (
Iberian Chelis species show how DNA barcodes divergences are often correlated with differences in morphology which can easily be overlooked or disputed (
The strong morphological similarity of all specimens in taxa with BIN splits further supports their recent separation although most cases may represent cryptic species complexes. In such cases, taxonomic decisions are often subjective and depend on the choice of species delimitation models, application of species concepts and taxonomic principles, especially when allopatric populations are involved (
Conclusions
1. Sequences for the barcode region of the mitochondrial COI gene from 271 specimens (160 species) of Erebidae moths (Insecta: Lepidoptera) from the Iberian Peninsula were gathered, showing a mean interspecific distance of 12.1%, while the mean nearest neighbour divergence was 6.4%.
2. All 160 species possessed diagnostic barcode sequences, but one pair of congeneric taxa were assigned to the same BIN. Intraspecific sequence divergences higher than 1.5% were detected in four species which likely represent species complexes.
3. This study reinforces the effectiveness of DNA barcoding as a tool for monitoring biodiversity in particular geographical areas and the strong correspondence between sequence clusters delineated by BINs and species recognized through detailed taxonomic analysis.
Acknowledgements
We are very grateful to the staff at the Canadian Centre for DNA Barcoding for sequence analysis and to many other colleagues involved in the Barcode of Life project (Centre for Biodiversity Genomics, Guelph, Canada) that contributed to this study. We are particularly grateful to Peter Huemer, Rodolphe Rougerie and Enrique Murria for granting access to their projects on BOLD; to Yerai Monasterio (La Rioja), Carlos Antonietty (Seville), Ramón Macià (Barcelona), Carmelo Abad, Aquilino Albaladejo, José de la Calle, Francisco Lencina and José Luis Palacios (Murcia), Pablo Valero and Maite Mójica (Alicante), Félix J. González-Estébanez (León) and Joseph de Freina (Munich) for loaning specimens and valuable comments. Thanks are also due to Pilar de la Rúa, Carlos Ruiz, Alejandro López, Ana Isabel Asensio and Carmelo Andújar for their comments, suggestions and technical support.
This study was supported by the project on insect barcoding CGL2009-10906 of the Spanish Ministry of Research and Science and by Regional Excellence 19908-GERM-15 project of the Fundación Seneca (Regional Government of Murcia, Spain). DNA sequencing was supported by Genome Canada through Ontario Genomics in the framework of the iBOL program. Collecting permits were issued by every Regional Administration and support was provided by environmental staff in each of them.
Hosting institution
Universidad de Murcia, Murcia, Spain
Ethics and security
No ethical principles were violated when performing this study.
Author contributions
A.O., J.S., and P.H. obtained funding. A.O., R.R., J.G., M.G. and A.H. collected the samples. A.O., R.R. and A.H. conceived and designed the experiments. A.O., R.R. and A.H. analysed the data. A.O., R.R., A.H. and P.H. wrote the manuscript. J.G., M.G. and J.S. contributed (additions/corrections) to the manuscrpt.
Conflicts of interest
The authors declare no conflict of interests concerning this study.
References
- DNA barcodes for biosecurity: invasive species identification.Philosophical Transactions of the Royal Society B: Biological Sciences360(1462):1813‑1823. https://doi.org/10.1098/rstb.2005.1713
- Comparative evidence for the evolution of genitalia by sexual selection.Nature393:784‑786. https://doi.org/10.1038/31689
- Balleto E, Casale A (1991) Mediterranean insect conservation. In: Collins NM, Thomas JA (Eds) The Conservation of Insects and their Habitats.Academic Press,London,121-142pp. https://doi.org/10.1016/b978-0-12-181370-3.50012-9
- DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae).Canadian Journal of Forest Research36(2):337‑350. https://doi.org/10.1139/x05-276
- DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica.Proceedings of the National Academy of Sciences105:6350‑6355. https://doi.org/10.1073/pnas.0712181105
- A comprehensive DNA barcode library for the looper moths (Lepidoptera: Geometridae) of British Columbia, Canada.PLoS ONE6(3):e18290. https://doi.org/10.1371/journal.pone.0018290
- Towards a global barcode library for Lymantria (Lepidoptera: Lymantriinae) tussock moths of biosecurity concern.PLoS ONE5(12):e14280. https://doi.org/10.1371/journal.pone.0014280
- deWaard JR, Ivanova NV, Hajibabaei M, Hebert PDN (2008) Assembling DNA barcodes: Analytical Protocols. In: Martin C (Ed.) Methods in Molecular Biology: Environmental Genetics.Humana Press,Totowa, NJ,275-293pp.
- Complete DNA barcode reference library for a country's butterfly fauna reveals high performance for temperate Europe.Proceedings of the Royal Society B: Biological Sciences278:347‑355. https://doi.org/10.1098/rspb.2010.1089
- DNA barcode reference library for Iberian butterflies enables a continental-scale preview of potential cryptic diversity.Scientific Reports5(12395):1‑12. https://doi.org/10.1038/srep12395
- Sexual selection and animal genitalia.Harvard University Press,Cambridge, MA,244pp.
- A comprehensive DNA sequence library is essential for identification with DNA barcodes.Molecular Phylogenetics and Evolution43(2):530‑542. https://doi.org/10.1016/j.ympev.2006.11.021
- Fauna Europaea: Erebidae. Fauna Europaea version 2.5. https://fauna-eu.org/. Accessed on: 2017-7-01.
- Die Bombyces und Sphinges der Westpalaerktis (Insecta, Lepidoptera) Band 1.Forschung & Wissenschaft,München, Germany,708pp. [InGerman].
- Origin of intra-individual variation in PCR-amplified mitochondrial cytochrome oxidase I of Thrips tabaci (Thysanoptera: Thripidae): mitochondrial heteroplasmy or nuclear integration?Hereditas140:92‑98. https://doi.org/10.1111/j.1601-5223.2004.01748.x
- Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent Approach: a revised method and evaluation on simulated data sets.Systematic Biology62(5):707‑724. https://doi.org/10.1093/sysbio/syt033
- Gómez A, Lunt D (2007) Refugia within refugia: atterns of phylogeographic concordance in the Iberian Peninsula. In: Weiss S, Ferrand N (Eds) Phylogeography of Southern European Refugia.Springer,Dordrecht, Netherlands,155-188pp. https://doi.org/10.1007/1-4020-4904-8_5
- DNA barcodes distinguish species of tropical Lepidoptera.Proceedings of the National Academy of Sciences103:968‑971. https://doi.org/10.1073/pnas.0510466103
- An integrative taxonomic approach to resolving some difficult questions in the Larentiinae of the Mediterranean region (Lepidoptera, Geometridae).Mitteilungen der Münchner Entomologischen Gesellschaft101:73‑97.
- DNA barcoding the geometrid fauna of Bavaria (Lepidoptera): successes, surprises, and questions.PLoS ONE6(2):e17134. https://doi.org/10.1371/journal.pone.0017134
- The geometrid moths of Ethiopia I: tribes Pseudoterpnini and Comibaenini (Lepidoptera: Geometridae, Geometrinae).Zootaxa3768:460‑468. https://doi.org/10.11646/zootaxa.3768.4.4
- The Geometrinae of Ethiopia II: Tribus Hemistolini, genus Prasinocyma (Lepidoptera: Geometridae, Geometrinae).Zootaxa4065(1):1‑63. https://doi.org/10.11646/zootaxa.4065.1.1
- Hypobapta tachyhalotaria n. sp. from Tasmania - an example of a new species revealed by DNA barcoding (Lepidoptera, Geometridae).Spixiana32:237‑242.
- Now DNA-barcoded: the butterflies and larger moths of Germany.Spixiana34:47‑58.
- Revision of the Australian Oenochroma vinaria Guenée, 1858 species-complex (Lepidoptera: Geometridae, Oenochrominae): DNA barcoding reveals cryptic diversity and assesses status of type specimen without dissection.Zootaxa2239:1‑21.
- Calibrating the taxonomy of a megadiverse insect family: 3000 DNA barcodes from geometrid type specimens (Lepidoptera, Geometridae).Genome59(9):671‑684. https://doi.org/10.1139/gen-2015-0197
- Genetic patterns in European geometrid moths revealed by the Barcode Index Number (BIN) system.PloS ONE8(12):e84518. https://doi.org/10.1371/journal.pone.0084518
- DNA barcodes for 1/1000 of the animal kingdom.Biology Letters6(3):359‑362. https://doi.org/10.1098/rsbl.2009.0848
- Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences270(1512):313‑321. https://doi.org/10.1098/rspb.2002.2218
- Hewitt G (2011) Mediterranean peninsulas: the evolution of hotspots. In: Zachos FE, Habel JC (Eds) Biodiversity Hotspots.Springer-Verlag,Berlin, Germany,123-147pp. https://doi.org/10.1007/978-3-642-20992-5_7
- Barcoding Lepidoptera of the Alps: the search for cryptic diversity.Barcode Bulletin3:4.
- Taxonomy of spatially disjunct alpine Teleiopsis albifemorella s. lat. (Lepidoptera: Gelechiidae) revealed by molecular data and morphology - how many species are there?Zootaxa3580:1‑23.
- Testing DNA barcode performance in 1000 species of European Lepidoptera: large geographic distances have small genetic impacts.PLoS ONE9(12):e115774. https://doi.org/10.1371/journal.pone.0115774
- A revised molecular phylogeny of the globally distributed hawkmoth genus Hyles (Lepidoptera: Sphingidae), based on mitochondrial and nuclear DNA sequences.Molecular Phylogenetics and Evolution52:852‑865. https://doi.org/10.1016/j.ympev.2009.05.023
- An inexpensive, automation-friendly protocol for recovering high-quality DNA.Molecular Ecology Notes6:998‑1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
- Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding.Philosophical Transactions of the Royal Society B: Biological Sciences360(1462):1835‑1845. https://doi.org/10.1098/rstb.2005.1715
- Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity.Molecular Ecology Resources9 Suppl s1:1‑26. https://doi.org/10.1111/j.1755-0998.2009.02628.x
- Konstantinov A, Korotyaev B, Volkovitsh M (2009) Insect biodiversity in the Palearctic Region. In: Foottit R, Adler P (Eds) Insect Biodiversity.Blackwell,Oxford, UK,107-162pp. https://doi.org/10.1002/9781444308211.ch7
- Moths of Europe, vol. 1: Saturnids, Lasiocampids, Hawkmoths, Tiger Moths,...NAP Editions,Verrières-le-Buisson, France,400pp.
- Identifying species of moths (Lepidoptera) from Baihua Mountain, Beijing, China, using DNA barcodes.Ecology and Evolution4(12):2472‑2487. https://doi.org/10.1002/ece3.1110
- DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification.Molecular Ecology Resources9(5):1302‑1310. https://doi.org/10.1111/j.1755-0998.2009.02577.x
- Detection and genetic diversity of a heliothine invader (Lepidoptera: Noctuidae) from north and northeast of Brazil.Journal of Economic Entomology107(3):970‑980. https://doi.org/10.1603/EC13403
- Advancing taxonomy and bioinventories with DNA barcodes.Philosophical Transactions of the Royal Society B: Biological Sciences371(1702):20150339. https://doi.org/10.1098/rstb.2015.0339
- Mooney HA (1988) Lessons from Mediterranean-climate regions. In: Wilson EO (Ed.) Biodiversity.National Academic Press,Washington, DC, USA,157-165pp.
- Allopatry as a Gordian knot for taxonomists: patterns of DNA barcode divergence in Arctic-Alpine Lepidoptera.PLoS ONE7:e47214. https://doi.org/10.1371/journal.pone.0047214
- Biodiversity hotspots for conservation priorities.Nature403:853‑858. https://doi.org/10.1038/35002501
- Use of DNA barcodes to identify invasive armyworm Spodoptera species in Florida.Journal of Insect Science11(1):154. https://doi.org/10.1673/031.011.15401
- DNA barcoding of the leaf-mining moth subgenus Ectoedemia s. str. (Lepidoptera, Nepticulidae) with COI and EF1-a: two are better than one in recognizing cryptic species.Contributions to Zoology81:1‑24.
- Order Lepidoptera Linnaeus, 1758. In: Zhang, Z.-Q. (Ed.) Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness.Zootaxa3148:212‑221.
- Oosterbroek P (1994) Biodiversity of the Mediterranean region. In: Forey PI, Humphries CJ, Vane-Wright RI (Eds) Systematics and conservation evaluation. Systematics Association Special Volume No. 50.Clarendon Press,Oxford, UK,289-307pp.
- Integrated taxonomy, phylogeography and conservation in the genus Chelis Rambur, [1866] in the Iberian Peninsula (Lepidoptera: Erebidae: Arctiinae).Spixiana39:273‑286.
- Pogue M (2009) Biodiversity of Lepidoptera. In: Foottit RG, Adler PH (Eds) Insect Biodiversity.Blackwell Publishing,London, UK,325-355pp. https://doi.org/10.1002/9781444308211.ch13
- Sequence-based species delimitation for the DNA taxonomy of undescribed insects.Systematic Biology55:595‑609. https://doi.org/10.1080/10635150600852011
- ABGD, Automatic Barcode Gap Discovery for primary species delimitation.Molecular Ecology21:1864‑1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
- BOLD Barcode of Life Data System, version 3. http://www.boldsystems.org. Accessed on: 2017-7-01.
- BARCODING: bold: The Barcode of Life Data System (http: // www. barcodinglife. org).Molecular Ecology Notes7:355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- A DNA-based registry for all animal species: the Barcode Index Number (BIN) system.PLoS ONE8:e66213. https://doi.org/10.1371/journal.pone.0066213
- Australian Sphingidae – DNA Barcodes Challenge Current Species Boundaries and Distributions.PLoS ONE9:e101108. https://doi.org/10.1371/journal.pone.0101108
- The lock-and-key hypothesis: evolutionary and biosystematic interpretation of insect genitalia.Annual Review of Entomology34:231‑245. https://doi.org/10.1146/annurev.en.34.010189.001311
- Wolbachia and DNA barcoding insects: patterns, potential, and problems.PloS one7:e36514. https://doi.org/10.1371/journal.pone.0036514
- Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified.Proceedings of the National Academy of Sciences105(36):13486‑13491. https://doi.org/10.1073/pnas.0803076105
- Catálogo sistemático y sinonímico de los Lepidoptera de la Península Ibérica, de Ceuta, de Melilla y de las islas Azores, Baleares, Canarias, Madeira y Salvajes (Insecta: Lepidoptera).Suplemento de SHILAP Revista de Lepidopterología,1184pp.
- Building a DNA barcode reference library for the true butterflies (Lepidoptera) of Peninsula Malaysia: what about the subspecies?PloS one8:e79969. https://doi.org/10.1371/journal.pone.0079969
- Noctuidae Europaeae. Lymantrinae-Arctiinae, including Phylogeny and Check List of the Quadrifid Noctuoidea of Europe. Volume 13.Entomological Press,Soro, Denmark,448pp.
- Manual de identificación y guía de campo de los Árctidos de la Península Ibérica y Baleares.Argania editio,Barcelona, Spain,290pp.
- A transcontinental challenge-a test of DNA barcode performance for 1,541 species of Canadian Noctuoidea (Lepidoptera).PloS ONE9:e92797. https://doi.org/10.1371/journal.pone.0092797
Supplementary materials
List of species names, sample-IDs, process-IDs (from BOLD database), COI-5P bps, BINs, GenBank Accession numbers, collection country, and Institution storing vouchers. Abbreviations: BIN = Barcode Index Number. NST = Naturmuseum Suedtirol. PCEM = Private Collection of Enrique Murria Beltran. RCAH = Research Collection of Alfred Haslberger. RCBA-UMU = Research Collection Biologia Animal-Universidad de Murcia. RCBD = Research Collection of Bernard Dardenne. RCCZ = Research Collection of Christian Zehentner. RCPL = Research Collection of Peter Lichtmannecker. RCRS = Research Collection of Ralph Sturm. RCTG = Research Collection of Theo Gruenewald. TLF = Tiroler Landesmuseum Ferdinandeum. ZSM = Zoologische Staatssammlung Muenchen.
Systematic list of subfamilies and species, barcode gap analysis (Mean and Maximum intraspecific variation and distance to nearest neighbor NN) for 160 Iberian species in the all European Erebidae
List of 16 Iberian taxa without a BIN assignment (awaiting DNA barcoding); a species with short sequences are marked with an *.
Genetic similarities visualized in a Neighbor Joining tree