|
Biodiversity Data Journal :
Research Article
|
|
Corresponding author: Nicholas C Goltz (nick.goltz@uconn.edu)
Academic editor: Matthew Yoder
Received: 22 May 2019 | Accepted: 29 Oct 2019 | Published: 17 Jan 2020
© 2020 Nicholas Goltz, Jessica Awad, Matthew Moore, Elijah Talamas
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Goltz NC, Awad J, Moore MR, Talamas EJ (2020) A fortuitous find: a unique haplotype of Ooencyrtus nezarae Ishii (Encyrtidae: Encyrtinae) discovered in Florida. Biodiversity Data Journal 8: e36440. https://doi.org/10.3897/BDJ.8.e36440
|
|
Abstract
The adventive arrival of biological control agents circumvents the regulatory process by introducing exotic species to control invasive pests and is generally followed by post hoc risk evaluation. The bean plataspid, Megacopta cribraria (Fabricius) (Hemiptera: Plataspidae), is an invasive pest of leguminous crops in the south-eastern United States that was eventually followed by two parasitoid wasps from its range in the eastern hemisphere, Paratelenomus saccharalis (Dodd) (Scelionidae) and Ooencyrtus nezarae Ishii (Encyrtidae). In North Central Florida, sentinel egg masses, intended to capture Paratelenomus saccharalis, instead yielded Ooencyrtus nezarae, which was previously known only from Alabama (Ademokoya et al. 2018). Two generations of O. nezarae were subsequently reared in the laboratory. COI sequences from the Florida population of O. nezarae differed by 1.3% from the Alabama population and the presence of a different haplotype suggests the possibility of a separate introduction. Laboratory parasitism rates, sex ratios, morphology, molecular diagnosis and implications for agriculture are discussed.
Keywords
Megacopta cribraria, Paratelenomus saccharalis, biological control, fortuitous biological control, species delimitation, DNA barcoding
Introduction
The term “fortuitous biological control” was coined by
This phenomenon has received renewed attention in recent years, due in part to the high-profile case of the samurai wasp, Trissolcus japonicus (Ashmead) (Hymenoptera: Scelionidae) (
Pentatomoid eggs, like the scale insects studied by
We recently discovered wild O. nezarae in North Central Florida. The publication of COI barcodes from the Alabama population of O. nezarae facilitated the identification of the parasitoids found in Florida. We here present rearing data, including observations of parasitism and emergence rates, morphological description and COI barcoding results for this population of O. nezarae.
Material and methods
Collection and rearing
On June 19, 2018, a collection trip was undertaken to replenish the colony of Paratelenomus saccharalis at the Florida Department of Agriculture and Consumer services – Division of Plant Industry (FDACS-DPI) in Gainesville, Florida. A patch of kudzu, Pueraria montana var. lobata (Lour.) Merr. (Fabaceae) located in Alachua County, Florida (
Of these M. cribraria egg masses, 47 were identified as having an irregular, ash-grey colouration, a common sign of parasitism (
Parasitism and emergence
Thirty Ooencyrtus nezarae adults were separated into three glass vials (V = 31 cm3 ; 10 parasitoids in each with an unknown sex ratio), each containing one M. cribraria egg mass and a honey-soaked strip of paper. Egg masses were obtained from captive colonies and stored in a freezer for 6 – 14 months before exposure. These vials were then sealed and maintained at 25 – 27°C, 60 – 78% humidity with a 16L:8D light cycle. The remaining parasitoids in the growth chamber were provided with four previously frozen, lab-reared M. cribraria egg masses and were maintained under the conditions described above.
Adult O. nezarae emergence in the glass vials was first observed 14 days later and continued for several more days. Once it was determined that adult parasitoids were unlikely to continue emerging, the 122 adults that emerged were individually placed in separate gelatine capsules and sorted by sex, resulting in 36 males and 86 females. Of these individuals, 32 males and 72 females were released into the eight glass vials, with each vial containing four males and nine females. Each new glass vial was provided with a new, previously frozen, lab-reared M. cribraria egg mass, to further evaluate rates of parasitism and suitability for rearing. The remaining adults were returned to the plexiglass growth chamber to be used as a maintenance colony.
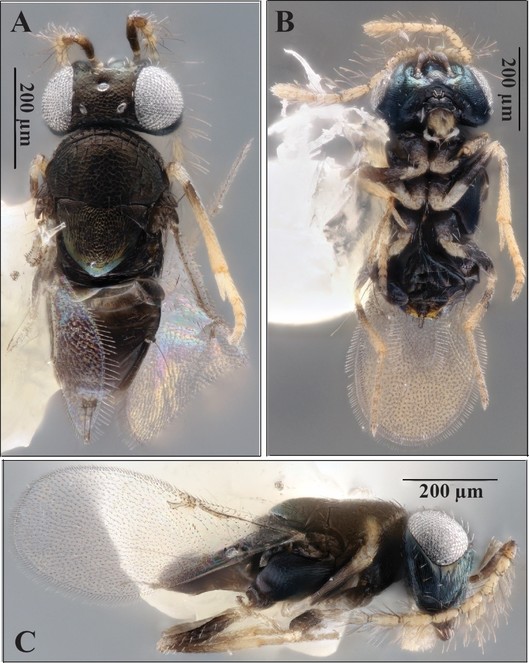
Morphological identification
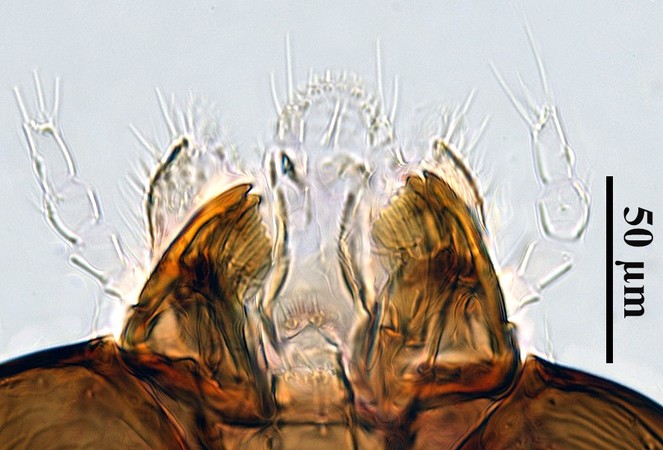
Specimens were point-mounted and deposited in the Florida State Collection of Arthropods (FDACS-DPI, E2018-4411-1). Two specimens (one male and one female) were cleared in potassium hydroxide and slide-mounted in Canada balsam, according to a protocol modified from
DNA extraction, PCR and sequence assembly
Six O. nezarae individuals from the DPI colony were selected for DNA extraction and COI barcoding. Samples were collected directly into 70% EtOH and were air-dried two hours before proceeding to DNA extraction. DNA extractions of entire individuals were performed using DNeasy Blood and Tissue Kits (Qiagen). Extraction protocols followed the manufacturer’s recommendations with one minor adjustment: samples were not incubated for 10 minutes following tissue lysis and addition of Buffer AL. DNA extracts were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific). At least 20 ng of template DNA was used per PCR.
The 5’-COI barcode region was PCR-amplified using the primers LEP-F1 and LEP-R1 (
Datamining, alignments and clustering analyses
To evaluate our new sequences in the context of the subfamily, all 5’-COI barcodes for Ooencyrtus and Encyrtinae were mined from the Barcode of Life Database (BOLD) (
Sequences were aligned using ClustalW with default settings (
Phylogenetic tree searches and species delimitation analyses
The mPTP species delimitation programme requires a rooted, binary Newick tree for input (
Delimitations computed in the mPTP programme used the minimum branch length threshold calculated with the “--minbr_auto” option. Heuristic maximum likelihood species delimitations were performed with a single coalescent rate averaged over all species. The confidence of the delimitations was assessed using the Markov Chain Monte Carlo (MCMC) sampling method. MCMC parameters included two independent runs with each run starting with the most likely delimitation, 100,000,000 MCMC iterations, and sampling of log-likelihoods every 10,000 MCMC iterations. The convergence of independent runs was evaluated by examining the sampled log-likelihood plots. The two runs converged within the first 1% of MCMC iterations.
Species delimitations were also produced using the server version of Automatic Barcode Gap Discovery (ABGD) (
Results
Parasitism and emergence
The three previously frozen M. cribraria egg masses used to rear the first generation comprised 235 eggs. Of these, 65 were deemed unviable because they had desiccated prior to being frozen or during the freezing process, resulting in 170 available eggs. Of the available eggs, 122 individuals emerged: 36 males and 86 females. The second generation provided a total of 751 eggs, with 521 eggs deemed viable. Of the viable eggs, 345 were parasitised (66.2%) and 314 adult O. nezarae emerged: 119 males and 195 females. Rates of parasitism and emergence for each of the vials are provided in Table
Rates of parasitism and emergence in the second generation Ooencyrtus nezarae.
|
Vial |
Available Host Eggs |
Number Parasitised |
Number Emerged |
Rate of Parasitism |
Proportion Female |
|
1 |
94 |
59 |
52 |
62.8% |
0.62 |
|
2 |
78 |
55 |
54 |
70.5% |
0.61 |
|
3 |
78 |
50 |
37 |
64.1% |
0.43 |
|
4 |
69 |
54 |
52 |
78.3% |
0.64 |
|
5 |
41 |
26 |
22 |
63.4% |
0.64 |
|
6 |
34 |
14 |
13 |
41.2% |
0.69 |
|
7 |
66 |
43 |
42 |
65.2% |
0.69 |
|
8 |
61 |
44 |
42 |
72.1% |
0.67 |
|
Total |
521 |
345 |
314 |
66.2% |
0.62 |
Morphological identification
Laboratory reared specimens were compared to descriptions in
There is some disagreement between
Clustering analyses
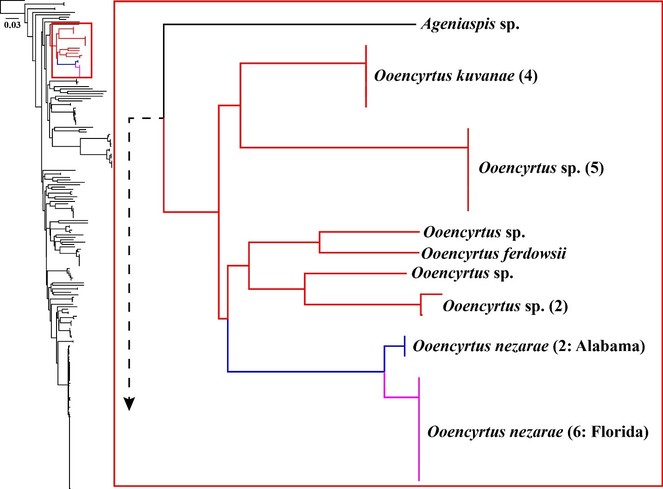
Neighbour-joining analysis excluded all positions with ambiguous data, leaving a total of 487 positions for inclusion (out of 562 possible positions) and recovered an Ooencyrtus group comprised of eight clusters (Fig.
Kimura 2-parameter neighbour-joining tree showing the clustering of similar sequences in the Encyrtinae 5’-COI barcode dataset. Red branches indicate the Ooencyrtus cluster. Blue and fuchsia branches indicate O. nezarae from Alabama and Florida, respectively. Numbers in parentheses after the taxon name indicate how many sequences are represented in that cluster.
Phylogenetic tree searches and species delimitation analyses
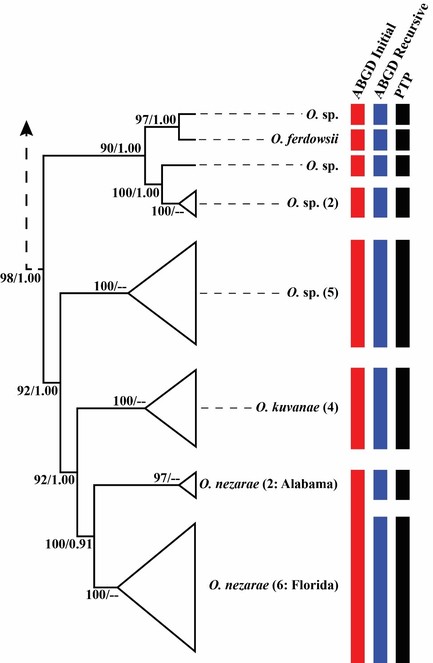
W-IQ-TREE analyses found the most likely tree with a log likelihood score of -12138.342. ML bootstrap support for the recovered Ooencyrtus clade was strong (98%; Fig.
Transformed Ooencyrtus branch from the most likely tree for the Encyrtinae COI dataset. Each coloured bar represents an OTU delimited by the ABGD (initial and recursive partitions) and PTP procedures. Support values are ML bootstraps and the fraction of MCMC sampled delimitations in which a node was part of the speciation process in the PTP procedure, respectively. Numbers in parentheses after the taxon name indicate how many sequences are represented in that OTU.
In ABGD, all tested minimum relative gap widths returned initial delimitations of 68 species from the Encyrtinae dataset at P = 0.01. In this same range of minimum relative gap widths, ABGD returned recursive delimitations of 71 or 72 from the encyrtine dataset at P = 0.01. Across all tests, the initial delimitations recognised seven Ooencyrtus species and lumped O. nezarae from Alabama and Florida into a single OTU (Fig.
Discussion
Our testing indicates that the Florida O. nezarae COI sequences are 1.3% different from the Alabama O. nezarae sequences. This was the largest infraspecific sequence divergence observed amongst the Ooencyrtus species in this study. PTP and ABGD recursive delimitations separated the Florida O. nezarae into distinct OTUs due to this degree of sequence divergence. Based on morphological data indicating that the Alabama and Florida O. nezarae are conspecific, inconsistency between molecular species delimitation methods and lack of statistical support for the PTP delimitation, we consider the divergent Florida O. nezarae COI sequences to represent a distinct haplotype rather than evidence of a separate species. Ooencyrtus nezarae is a widely distributed species occurring in China, Japan, Thailand, South Korea, Brazil and now the south-eastern United States (
Implications for Florida agriculture merit further investigation. Ooencyrtus nezarae has a wide host range, including at least four hemipteran families (
Future research may investigate the performance of O. nezarae on other pentatomoid host species of interest to Florida agriculture. Morphometrics of parasitoids reared from different sizes and species of eggs are expected to reveal allometric scaling. Due to this, antennal morphometrics are not recommended as diagnostic characters for this species, although they may have some utility in determining the host origin of specimens.
Competition between egg parasitoid species has the potential to impact populations of introduced and native biological control agents. For example, egg parasitoids, such as Trissolcus species, are known to exhibit intra-guild competition, both in the form of exploitative competition when parasitoids of another genus are present and in the form of interference competition when multiple Trissolcus species are present (
At the time of writing, fifteen generations of O. nezarae have been reared successfully in captivity using previously frozen, lab-reared Megacopta cribraria eggs, collected 6 – 14 months prior to use. This is the first recorded instance of O. nezarae being reared in captivity using previously frozen eggs or from eggs kept in storage, refrigerated or frozen, for more than 200 days (
Conclusions
Ooencyrtus nezarae is an interesting new arrival to Florida; one that merits further study. Through rearing, we have observed that it is a hardy, generalist egg parasitoid well-adapted to a subtropical climate. The ease by which it can be reared suggests it may be a suitable candidate for future biological control projects requiring a generalist parasitoid, such as augmentative biological control programmes for growers dealing with hemipteran pests.
Despite possible beneficial applications this insect may have, however, the fact remains that it is not indigenous to the United States. Additional research is necessary to determine if O. nezarae may negatively impact populations of native egg parasitoids or beneficial hemipterans. Additionally, native populations should be sampled and sequenced to elucidate why the Florida population is of a different haplotype than the previously-documented Alabama population. Determining where these respective non-indigenous populations originated may provide novel information regarding the pathways by which invasive organisms arrive in the south-eastern United States.
Acknowledgements
We thank the Florida Department of Agriculture and Consumer Services – Division of Plant Industry for their support on this contribution. We are very grateful to Amy Howe, Eric Rohrig, Cheryl Roberts and George Schneider (FDACS-DPI) for their advice and scientific support. This work would not have been possible without Kylie Lennon (University of Florida), who imaged the wasp mandibles; Andy Boring and Sedonia Steininger (FDACS-DPI), who reviewed our manuscript and provided constructive comments; and librarian Jeff Eby (FDACS-DPI), who was instrumental in finding Japanese literature.
References
- The first record of Ooencyrtus nezarae (Hymenoptera: Encyrtidae) on kudzu bug (Hemiptera: Plataspidae) in North America.Journal of Insect Science18:1‑7.
- Biological attributes of Ooencyrtus nezarae Ishii (Hymenoptera: Encyrtidae) reared on refrigerated eggs of Riptortus pedestris (=clavatus) Fabricius (Hemiptera: Alydidae).Journal of Asia-Pacific Entomology13:139‑143. https://doi.org/10.1016/j.aspen.2010.01.004
- DeBach P (1971) Fortuitous biological control from ecesis of natural enemies. In: Asahinas S, Gressitt J, Hidaka Z, Nishida T, Nomura K (Eds) Entomological Essays to Commemorate the Retirement of Professor K. Yasumatsu.Hokuryucan Publishing Company,Tokyo. https://doi.org/10.1007/978-1-4615-6531-4_7
- Biological control by natural enemies.Second edition.Cambridge University Press,New York/Port Chester/Melbourne/Sydney,440pp.
- Trissolcus hyalinipennis Rajmohana & Narendran (Hymenoptera, Scelionidae), a parasitoid of Bagrada hilaris (Burmeister) (Hemiptera, Pentatomidae), emerges in North America.Journal of Hymenoptera Research65:111‑130. https://doi.org/10.3897/jhr.65.25620
- Discovery of Paratelenomus saccharalis (Dodd) (Hymenoptera: Platygastridae), an egg parasitoid of Megacopta cribraria F. (Hemiptera: Plataspidae) in its North American range.Journal of Entomological Science48:355‑359. https://doi.org/10.18474/0749-8004-48.4.355
- Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator.PNAS101:14812‑14817. https://doi.org/10.1073/pnas.0406166101
- UFBoot2: improving the ultrafast bootstrap approximation.Molecular Biology and Evolution35:518‑522. https://doi.org/10.1093/molbev/msx281
- The Encyrtinae of Japan.Bulletin of the Imperial Agricultural Experiment Station of Japan3:79‑160.
- ModelFinder: fast model selection for accurate phylogenetic estimates.Nature Methods14:587‑589. https://doi.org/10.1038/nmeth.4285
- Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo.Bioinformatics33:1630‑1638.
- A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences.Journal of Molecular Evolution16:111‑120. https://doi.org/10.1007/BF01731581
- Integrated control of soybean stink bugs in the Cerrados.Japan Agricultural Research Quarterly20:231‑236.
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets.Molecular Biology and Evolution33:1870‑1874. https://doi.org/10.1093/molbev/msw054
- First record of Paratelenomus saccharalis (Hymenoptera: Platygastridae) on kudzu bug Megacopta cribraria (Heteroptera: Plataspidae) in Florida.Florida Entomologist98:1250‑1251. https://doi.org/10.1653/024.098.0438
- Discovery of an exotic egg parasitoid of the Brown Marmorated Stink Bug, Halyomorpha halys (Stål) in the Pacific Northwest.Proceedings of the Entomological Society of Washington118(3):466‑470. https://doi.org/10.4289/0013-8797.118.3.466
- A molecular phylogeny of the Chalcidoidea (Hymenoptera).PLoS One6(11):e27023. https://doi.org/10.1371/journal.pone.0027023
- Universal Chalcidoidea Database. http://www.nhm.ac.uk/chalcidoids/
- Collecting and preserving chalcid wasps (Hymenoptera: Chalcidoidea).Journal of Natural History16:315‑334. https://doi.org/10.1080/00222938200770261
- Algorithmic single-locus species delimitation: effects of sampling effort, variation and nonmonophyly in four methods and 1870 species of beetles.Molecular Ecology Resources17:393‑404. https://doi.org/10.1111/1755-0998.12557
- ABGD, Automatic Barcode Gap Discovery for primary species delimitation.Molecular Ecology21:1864‑1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
- FigTree v1.4.http://tree.bio.ed.ac.uk/software/figtree/
- BOLD: The Barcode of Life data system.Molecular Ecology Notes7:355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- Control freaks.Science361(6402):542‑545. https://doi.org/10.1126/science.361.6402.542
- First discovery of Trissolcus japonicus in Europe.Journal of Pest Sciencehttps://doi.org/10.1007/s10340-018-1061-2
- Inter and intra-guild interactions in egg parasitoid species of the soybean stink bug complex.Pesquisa Agropecuária Brasileira37:1541‑1549. https://doi.org/10.1590/S0100-204X2002001100004
- Trissolcus japonicus (Ashmead) (Hymenoptera, Scelionidae) emerges in North America.Journal of Hymenoptera Research43:119‑128. https://doi.org/10.3897/JHR.43.4661
- CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matric choice.Nucleic Acids Research22:4673‑4680. https://doi.org/10.1093/nar/22.22.4673
- W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis.Nucleic Acids Research44:232‑235. https://doi.org/10.1093/nar/gkw256
- A taxonomic study of Chinese species of Ooencyrtus (Insecta: Hymenoptera: Encyrtidae).Zoological Studies44:347‑360.