|
Biodiversity Data Journal :
Taxonomic paper
|
Unveiling of a cryptic Dicranomyia (Idiopyga) from northern Finland using integrative approach (Diptera, Limoniidae)
|
Corresponding author:
Academic editor: Pierfilippo Cerretti
Received: 11 Nov 2014 | Accepted: 28 Nov 2014 | Published: 03 Dec 2014
© 2014 Jukka Salmela, Kari Kaunisto, Varpu Vahtera
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Salmela J, Kaunisto K, Vahtera V (2014) Unveiling of a cryptic Dicranomyia (Idiopyga) from northern Finland using integrative approach (Diptera, Limoniidae). Biodiversity Data Journal 2: e4238. https://doi.org/10.3897/BDJ.2.e4238
|
|
Abstract
The subgenus Idiopyga Savchenko, 1987 is a northern hemisphere group of short-palped crane flies (Diptera, Limoniidae). In the current article we describe a new species, Dicranomyia (I.) boreobaltica Salmela sp.n., and redescribe the male and female post-abdomen of a closely related species, D. (I.) intricata Alexander. A standard DNA barcoding fragment of 5′ region of the cytochrome c oxidase I (COI) gene of the new species is presented, whilst the K2P minimum distances between the new species and 10 other species of the subgenus were found to range from 5.1 to 15.7 % (mean 11.2 %). Phylogenetic analyses (parsimony and maximum likelihood) based on COI sequences support the identity of the new species and its close relationship with D. (I.) intricata and D. (I.) esbeni (Nielsen). The new species is known from the northern Baltic area of Finland. The new species has been mostly collected from Baltic coastal meadows but an additional relict population is known from a calcareous rich fen that was estimated to have been at sea level circa 600-700 years ago. Dicranomyia (I.) intricata (syn. D. suecica Nielsen) is a Holarctic species, occurring in the north boreal and subarctic vegetation zones in Fennoscandia.
Keywords
Crane flies, DNA barcoding, Baltic coastal meadows, mires
Introduction
The subgenus Idiopyga (
Dicranomyia (I.) intricata was reported from Finland by Nieminen (
Materials and methods
The morphological terminology used here mainly follows
Layer photos were taken using an Olympus E520 digital camera, attached to an Olympus SZX16 stereomicroscope. Digital photos were captured using the programmes Deep Focus 3.1 and Quick PHOTO CAMERA 2.3. Layer photos were finally combined with the program Combine ZP.
A 658 bp fragment of mitochondrial protein-encoding cytochrome c oxidase subunit I (COI) was sequenced from a total of 22 Dicranomyia specimens and one Metalimnobia specimen. Legs or 2–3 abdominal segments of the specimens were placed in 96% ethanol in a 96-well lysis microplate and dispatched to the Canadian Centre for DNA Barcoding, Biodiversity Institute of Ontario where DNA was extracted and sequenced using standard protocols and primers (
Dicranomyia (Idiopyga) and outgroup specimens (Dicranomyia (D.) didyma (Meigen), D. (Numantia) fusca (Meigen), Metalimnobia (M.) charlesi Salmela & Starý) used in DNA barcoding (COI). Co-ordinates are given in WGS84 decimal format.
| species_sample ID | GenBank | year | country | locality | N | E |
| Dicranomyia danica_JES-20110422 | KP064166 | 2009 | Czech Republic | Hrabetice | 48.788 | 16.426 |
| Dicranomyia danica_JES-20110418 | KP064167 | 2009 | Czech Republic | Hrabetice | 48.788 | 16.426 |
| Dicranomyia esbeni_JES-20120182 | KP064169 | 2005 | Finland | Oulunsalo | 65.039 | 24.818 |
| Dicranomyia halterella_JES-20120184 | KP064171 | 2008 | Finland | Kankaanpää | 61.768 | 22.639 |
| Dicranomyia halterella_JES-20110097 | KP064172 | 2009 | Finland | Enontekiö | 68.636 | 22.784 |
| Dicranomyia intricata_JES-20120082 | KP064173 | 2009 | Finland | Kittilä | 67.639 | 25.427 |
| Dicranomyia intricata_JES-20110082 | KP064174 | 2009 | Finland | Enontekiö | 68.636 | 22.538 |
| Dicranomyia boreobaltica_JES-20120094 | KP064175 | 2005 | Finland | Oulunsalo | 64.906 | 25.376 |
| Dicranomyia klefbecki_JES-20120377 | KP064176 | 2011 | Finland | Eckerö | 60.253 | 19.541 |
| Dicranomyia lulensis_JES-20120081 | KP064177 | 2009 | Finland | Kemijärvi | 66.997 | 27.150 |
| Dicranomyia lulensis_JES-20110365 | KP064178 | 2007 | Finland | Enontekiö | 68.484 | 22.353 |
| Dicranomyia lulensis_JES-20110142 | KP064179 | 2009 | Finland | Enontekiö | 68.660 | 22.638 |
| Dicranomyia magnicauda_JES-20120119 | KP064180 | 2007 | Finland | Kittilä | 68.026 | 25.111 |
| Dicranomyia magnicauda_JES-20120080 | KP064181 | 2009 | Finland | Kemijärvi | 66.997 | 27.150 |
| Dicranomyia magnicauda_JES-20120122 | KP064182 | 2007 | Finland | Kittilä | 67.589 | 25.662 |
| Dicranomyia murina_JES-20120042 | KP064183 | 2009 | Finland | Sodankylä | 68.087 | 26.109 |
| Dicranomyia ponojensis_JES-20110117 | KP064184 | 2009 | Finland | Enontekiö | 68.636 | 22.538 |
| Dicranomyia ponojensis_JES-20120086 | KP064185 | 2007 | Finland | Suomussalmi | 65.230 | 28.170 |
| Dicranomyia stigmatica_JES-20120415 | KP064186 | 2006 | Finland | Ruovesi | 61.837 | 24.064 |
| Dicranomyia stigmatica_JES-20110360 | KP064187 | 2007 | Finland | Kittilä | 67.593 | 25.308 |
| Dicranomyia didyma_JES-20110098 | KP064168 | 2009 | Finland | Enontekiö | 68.636 | 22.784 |
| Dicranomyia fusca_JES-20110237 | KP064170 | 2008 | Finland | Nurmes | 63.786 | 29.350 |
| Metalimnobia charlesi_JES-20110381 | KP064165 | 2008 | Finland | Lieksa | 63.468 | 29.942 |
Phylogenetic analyses
Parsimony approach
Since the number of studied taxa was only 23, no heuristic methods were needed for the parsimony analysis. This allowed us to explore all possible evolutionary hypotheses for our data via explicit enumeration (branch and bound) analysis in TNT (Tree Analysis using New Technology) version 1.1 (
Maximum likelihood approach
Maximum likelihood analysis was conducted with RAxML ver. 8.0.22 (
Taxon treatments
Dicranomyia (Idiopyga) boreobaltica , sp. n.
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars ouluensis; verbatimLocality:Oulunsalo, Papinkari; verbatimLatitude:64.9060; verbatimLongitude:25.3764; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2005-8-11/10-8; habitat:Baltic coastal meadow; individualCount:1; sex:male; catalogNumber:JES-20120094; recordedBy:T. Nieminen; institutionCode:ZMUT
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars ouluensis; verbatimLocality:Hailuoto, Pökönnokka; verbatimLatitude:65.0790; verbatimLongitude:24.8883; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2005-8-11/10-8; habitat:Baltic coastal meadow; individualCount:1; sex:male; recordedBy:T. Nieminen; institutionCode:JSO
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars ouluensis; verbatimLocality:Hailuoto, Pökönnokka; verbatimLatitude:65.0790; verbatimLongitude:24.8883; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2005-8-11/10-8; habitat:Baltic coastal meadow; individualCount:1; sex:male; recordedBy:T. Nieminen; institutionCode:ZMUT
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars borealis; verbatimLocality:Tornio, Isonkummunjänkä Mire Conservation Area, Kusiaiskorpi; verbatimLatitude:65.8880; verbatimLongitude:24.4792; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2013-8-1/9-26; habitat:Rich fen, rusty spring; individualCount:1; sex:male; catalogNumber:DIPT-JS-2014-0248; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars borealis; verbatimLocality:Tornio, Isonkummunjänkä Mire Conservation Area, Kusiaiskorpi; verbatimLatitude:65.8880; verbatimLongitude:24.4792; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2013-8-1/9-26; habitat:Rich fen, rusty spring; individualCount:1; sex:female; catalogNumber:DIPT-JS-2014-0251; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars borealis; verbatimLocality:Tornio, Isonkummunjänkä Mire Conservation Area, Kusiaiskorpi; verbatimLatitude:65.8880; verbatimLongitude:24.4792; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2013-8-1/9-26; habitat:Rich fen, rusty spring; individualCount:1; sex:female; recordedBy:J. Salmela; institutionCode:ZMUT
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:boreobaltica; scientificNameAuthorship:Salmela; country:Finland; stateProvince:Ostrobothnia borealis pars borealis; verbatimLocality:Tornio, Isonkummunjänkä Mire Conservation Area, Kusiaiskorpi; verbatimLatitude:65.8880; verbatimLongitude:24.4792; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2013-8-1/9-26; habitat:Rich fen, rusty spring; individualCount:7; sex:4 females, 3 males; catalogNumber:DIPT-JS-2014-0114; recordedBy:J. Salmela; institutionCode:JES
Description
Dicranomyia (Idiopyga) intricata
Dicranomyia (Idiopyga) cf. intricata
Male. Head. Vertex dark brown, with short black setae. Rostrum light brown with a few short dark setae. Palpus 5-segmented; first palpomere very short, globular, 1.5 times wider than long; other palpomeres elongated, p2 length 140 µm, p3 100 µm, p4 100 µm and p5 120 µm. First palpomere with a long ventral seta, approximately 2 times longer than width of palpomere. Second and third palpomeres with 5 setae, arranged in the apical half of segments. Fourth palpomere bearing ca. 12 setae and p5 with 13-15 setae, most of these on the apices of the segments. Antennae 14-segmented, dark brown, segments bearing black setae mostly exceeding width of respective segment; setae straight on scape (ca. 10 setae) and pecidel (ca. 15 setae), straight or curved on flagellomeres (ca. 5 setae on each flagellomere). Scape cylindrical, length 200 µm, width 75 µm, pedicel wider apically than basally, length 115 µm, width 75 µm. Flagellomeres oval, longer than wide; f1 length 120 µm, width 65 µm, f2 length 8 µm, width 5 µm, f10 length 110 µm, width 40 µm. Thorax mainly dark brown. Prescutum dark brown, only small yellowish spots on hind lateral corners. Scutum dark brown with longitudinal yellow median line and yellow lateral spots near wing base. Mediotergite and anepisternum dark brown, mediotergite sometimes with narrow yellowish anterior margin. Laterotergite and anepimeron yellowish brown. Katepisternum bicolored: anterior half dark brown, posterior half yellowish brown. Fore coxa brown, mid and hind coxae yellowish brown. Femorae light brown or brown, tibiae and tarsi dark brown. Length of fore femora 4500 µm, tibia 5250 µm, t1 3500 µm, t2 1100 µm, t3 875 µm, t4 300 µm, t5 175 µm, claw 130 µm. Length of mid femora 5575 µm, tibia 5625 µm, t1 3200 µm, t2 1150 µm, t3 625 µm, t4 275 µm, t5 175 µm, claw 130 µm. Length of hind femora 5600 µm, tibia 5750 µm, t1 3050 µm, t2 1150, t3 650 µm, t4 275 µm, t5 178 µm, claw 130 µm. Halter grayish-brown. Wing clear, veins light brown - brown, pterostigma brown (Fig.
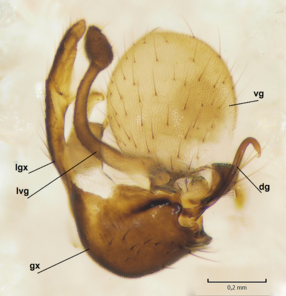
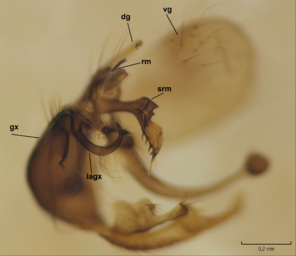
Dicranomyia (I.) boreobaltica Salmela sp.n. male hypopygium, gonocoxite and gonostylus. gx=gonocoxite, algx=appendage of ventromesal lobe of gonocoxite, lgx=ventromesal lobe of gonocoxite, dg=dorsal lobe of gonostylus, vg=ventral lobe of gonostylus.
b: Gonocoxite and gonostylus, outer/lateral view.
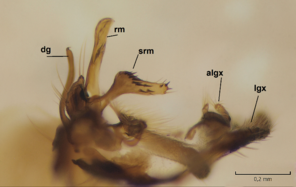
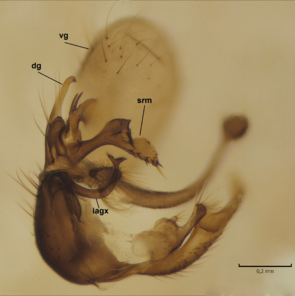
Dicranomyia (I.) boreobaltica Salmela sp.n., details of male hypopygium. vg=ventral lobe of gonostylus, dg=dorsal lobe of gonostylus, gx=gonocoxite, rm=rostral prolongation (rostrum) of ventral gonostyle, srm=subrostral prolongation of ventral gonostyle, iagx=inner appendage of gonocoxite, lgx=ventromesal lobe of gonocoxite, algx=appendage of ventromesal lobe of gonocoxite.
b: Gonostylus and gonocoxite, mesal view.
c: Gonostylus and gonocoxite, lateromesal view.
d: Gonostylus and gonocoxite, ventromesal view.
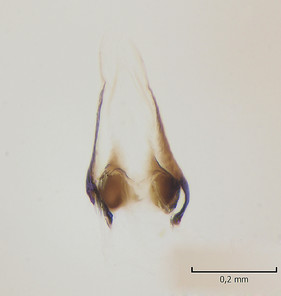
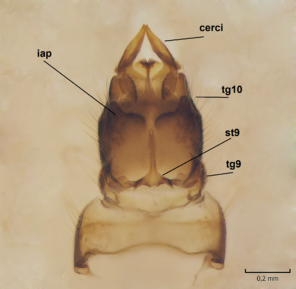
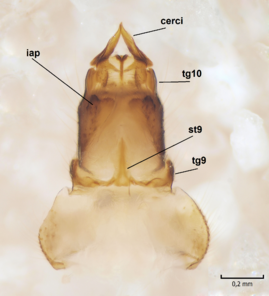
Female. In general, similar to male. Wing length 6.5 mm. Cerci short, ca. 240 µm in length. Infra-anal plate with a strong caudal peak (Fig.
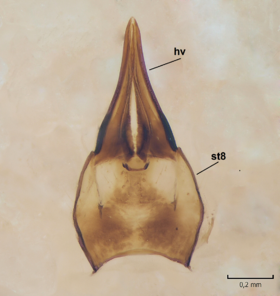
Dicranomyia (I.) boreobaltica Salmela sp.n., female, details of post-abdomen.
b: Hypogynial valves, inner/dorsal view. hv=hypogynial valves, st=sternite.
Dicranomya (I.) intricata Alexander, male hypopygium, gonocoxite and gonostylus. gx=gonocoxite, algx=appendage of ventromesal lobe of gonocoxite, lgx=ventromesal lobe of gonocoxite, dg=dorsal lobe of gonostylus, vg=ventral lobe of gonostylus, lvg=ventrobasal lobe of ventral gonostyle.
b: Gonocoxite and gonostylus, outer/lateral view.
Dicranomyia (I.) intricata Alexander, details of male hypopygium. vg=ventral lobe of gonostylus, dg=dorsal lobe of gonostylus, gx=gonocoxite, rm=rostral prolongation (rostrum) of ventral gonostyle, srm=subrostral prolongation of ventral gonostyle, iagx=inner appendage of gonocoxite, lgx=ventromesal lobe of gonocoxite, algx=appendage of ventromesal lobe of gonocoxite.
b: Gonocoxite and gonostylus, mesal view.
c: Gonostylus and gonocoxite, lateromesal view.
d: Gonostylus and gonocoxite, dorsomesal view.
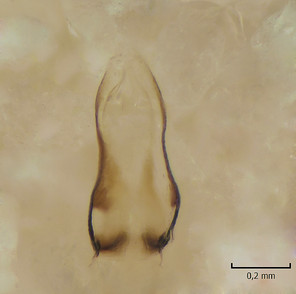
Dicranomyia (I.) intricata Alexander, female, details of postabdomen.
b: Hypogynial valves, inner/dorsal view. hv=hypogynial valves, st=sternite.
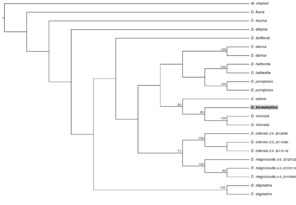
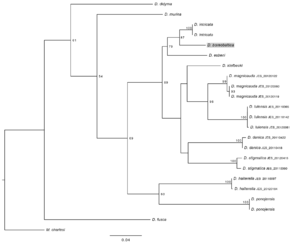
Phylogenetic trees of Dicranomyia (Idiopyga) species and three out-group species based on COI sequences (raw data available in Suppl. material
b: The optimal maximum likelihood tree (lnL= -3501.956818) with bootstrap support on nodes.
Diagnosis
Brownish, small species, very close to D. (I.) intricata. Ventrobasal lobe of ventral gonostylus sinuous, apex oval. Inner appendage of gonocoxite apically rounded. Rostral prolongantion of ventral apically rather narrow and subrostral prolongation simple, not bilobed, bearing dark stout spines. Female infra-anal plate with strong caudal peak.
Etymology
Boreo (borealis, Latin)= north, baltica (Latin)= referring to the Baltic Sea. The species is so far known from the northern Baltic area. The species name is deemed to be a latinized adjective in nominative singular.
Distribution
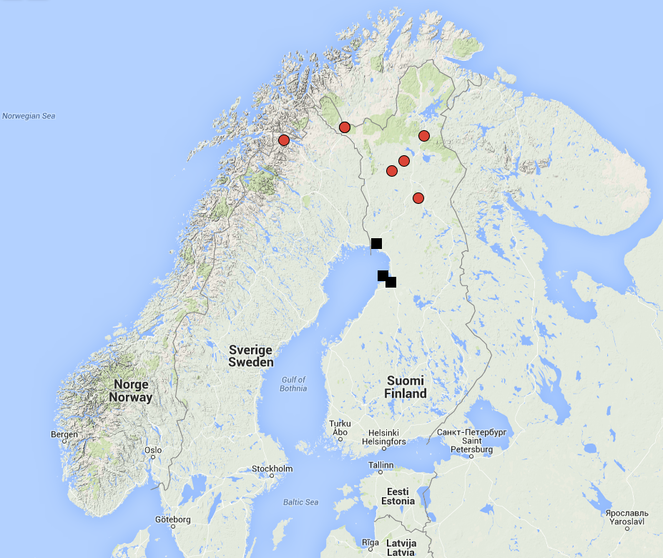
European, only known from Finland. The species is hitherto known from five separate localities; four of these are shore meadows in Oulunsalo and Hailuoto island (see
Ecology
The species is probably halophilous, occurring in Baltic coastal meadows characterised by vascular plants such as Phragmites australis, Lysimachia thyrsiflora, Eleocharis palustris, Carex halophila and C. paleacea (
Conservation
Due to its apparent rarity, that is, small area of occupancy and extent of occurrence, the species could most likely be assessed as a threatened species according to IUCN criteria. Habitats of this species are highly endangered, usually small and isolated. There are a total of ca. 4200 ha of Baltic coastal meadows along the Finnish coast, and all such habitat types are red-listed (
Taxon discussion
Based on morphology and COI sequence divergence, the new species is very closely related to the Holarctic species D. (I.) intricata. As already stated in the title of this article, the new species is cryptic, meaning that it is hard to distinguish from its sister species by morphological characters. Strictly speaking, cryptic species may mean taxa that are morphologically indisguishable (
External characters, such as wing venation and body coloration, between D. (I.) boreobaltica Salmela sp.n. and D. (I.) intricata are practically identical. The most important differences in male and female post-abdomen between the species are summarized in Table
Summary of the most important postabdominal differences between Dicranomyia (I.) boreobaltica Salmela sp.n. and D. (I.) intricata Alexander.
| D. (I.) boreobaltica | D. (I.) intricata |
| apex of iagx simple, not furcated (Fig. |
apex of iagx bifurcated (Fig. |
| apex of lgx angular (Fig. |
apex of lxg beak-like (Fig. |
| stalk of lvg rather wide, apex oval (Fig. |
stalk of lvg tapering apically, apex spherical (Fig. |
| apex of rm rounded, rather narrow (Fig. |
apex of rm pointed, rather wide (Fig. |
| srm simple, not bilobed (Fig. |
srm bilobed (Fig. |
| caudal margin of female infra-anal plate as in Fig. |
caudal margin of female infra-anal plate as in Fig. |
DNA barcode
Standard 5′ region (658 bp) of the cytochrome c oxidase I (COI) sequence of Dicranomyia (I.) boreobaltica Salmela sp.n. BOLD Sample ID JES-20120094, holotype specimen:
TACCTTATACTTTATTTTTGGAGCTTGAGCAGGAATAGTGGGAACTTCATTAAGTATTATTATTCGAGCAGAATTAGGACACCCAGGTGCATTAATTGGAGACGACCAGATTTATAATGTGGTAGTTACTGCCCATGCTTTTATTATAATTTTCTTTATAGTTATACCAATTATAATTGGAGGATTCGGTAATTGATTAGTTCCTTTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGAATACTTCCCCCTTCTTTAACTTTATTATTAGCTAGAAGCATAGTTGAAAACGGGGCAGGAACTGGCTGAACAGTATACCCTCCCCTTTCTTCTGGAATTGCCCATTCAGGGGCTTCTGTAGATTTAGCTATTTTTTCTCTTCACCTAGCAGGTATTTCTTCTATTTTAGGAGCTGTTAATTTTATTACAACTGTTATTAATATACGTTCAGCAGGAATTTCATTTGATCGAATACCATTATTTGTTTGATCAGTAGTAATTACTGCTATTTTATTGCTTTTATCACTTCCTGTTTTAGCCGGAGCTATTACAATATTATTAACAGATCGAAACTTAAATACTTCATTTTTTGATCCCGCAGGTGGAGGAGACCCTATTTTATATCAGCATTTATTT
Based on K2P (
Dicranomyia (Idiopyga) intricata
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:Alberta; verbatimLocality:Lesser Slave Lake; verbatimLatitude:55.35; verbatimLongitude:-115.09; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; eventDate:1924-8-1; individualCount:1; sex:male; recordedBy:O. Bryant; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:suecica; scientificNameAuthorship:Nielsen; country:Sweden; stateProvince:Abisko; verbatimLatitude:68.35; verbatimLongitude:18.79; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; eventDate:unknown; individualCount:1; sex:male; catalogNumber:855; recordedBy:H. Frantz; institutionCode:ZMUC
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:Alberta; verbatimLocality:Lesser Slave Lake, Grizzly mt.; minimumElevationInMeters:914; eventDate:1924-8-15; individualCount:1; sex:male; recordedBy:O. Bryant; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:Yukon; verbatimLocality:Dawson; minimumElevationInMeters:335; eventDate:1949-8-6; individualCount:1; sex:male; recordedBy:P.T. Bruggemann; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:British Columbia; verbatimLocality:Telegraph Creek; minimumElevationInMeters:335; eventDate:1960-8-28; individualCount:1; sex:male; recordedBy:W.W. Moss; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:British Columbia; verbatimLocality:Telegraph Creek, Sawmill Lake; eventDate:1960-8-18; individualCount:1; sex:male; recordedBy:W.W. Moss; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Canada; stateProvince:Northwest Territories; verbatimLocality:Aklavik; eventDate:1931-8-27; individualCount:1; sex:male; recordedBy:O. Bryant; institutionCode:USNM
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia kemensis pars occidentalis; verbatimLocality:Kittilä, Mustaoja-Nunaravuoma Mire Conservation Area, Mustaoja W; verbatimLatitude:67.6390; verbatimLongitude:25.4277; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2009-8-19; habitat:rich flark fen; individualCount:1; sex:female; catalogNumber:JES-20120082; recordedBy:J. Salmela; institutionCode:ZMUT
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia enontekiensis; verbatimLocality:Enontekiö, Tarvantovaara Wilderness Area, Tomuttirova W; verbatimLatitude:68.6369; verbatimLongitude:22.5381; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2009-8-26; habitat:swampy flark fen; individualCount:32; sex:29 male, 3 female; catalogNumber:DIPT-JS-2014-0336; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia enontekiensis; verbatimLocality:Enontekiö, Tarvantovaara Wilderness Area, Tomuttirova W; verbatimLatitude:68.6369; verbatimLongitude:22.5381; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2009-8-26; habitat:swampy flark fen; individualCount:1; sex:male; catalogNumber:JES-20110082; recordedBy:J. Salmela; institutionCode:ZMUT
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia enontekiensis; verbatimLocality:Enontekiö, Tarvantovaara Wilderness Area, Tomuttirova N; verbatimLatitude:68.6391; verbatimLongitude:22.5518; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2009-8-26; habitat:intermediate rich flark fen; individualCount:10; sex:5 male, 5 female; catalogNumber:DIPT-JS-2014-0337; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia kemensis pars orientalis; verbatimLocality:Sodankylä, Pomokaira-Tenniöaapa Mire Conservation Area, Syväkuru; verbatimLatitude:67.8718; verbatimLongitude:26.2126; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:Malaise trap; eventDate:2013-8-15/9-19; habitat:spring fen; individualCount:1; sex:male; catalogNumber:DIPT-JS-2014-0182; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Ostrobothnia borealis pars borealis; verbatimLocality:Kemijärvi, Salmiaavanhete; verbatimLatitude:66.9929; verbatimLongitude:27.0578; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2009-8-15; habitat:rich flark fen; individualCount:4; sex:1 male, 3 female; catalogNumber:DIPT-JS-2014-0338; recordedBy:J. Salmela; institutionCode:JES
-
genus: Dicranomyia; subgenus:Idiopyga; specificEpithet:intricata; scientificNameAuthorship:Alexander; country:Finland; stateProvince:Lapponia inariensis; verbatimLocality:Inari, Kaunispää; verbatimLatitude:68.4461; verbatimLongitude:27.4351; verbatimCoordinateSystem:decimal degrees; verbatimSRS:WGS84; samplingProtocol:sweep net; eventDate:2013-8-16; habitat:alpine wetland; individualCount:1; sex:1 female; catalogNumber:DIPT-JS-2014-0340; recordedBy:J. Salmela; institutionCode:JES
Description
Dicranomyia intricata
Limonia (Dicranomyia) suecica
Limonia (Dicranomyia) suecica
Dicranomyia (Idiopyga) intricata
The holotype specimen of D. (I.) intricata (Fig.
Male hypopygium. 9th tergite and proctiger as in Fig.
Female postabdomen. Cerci and hypogynial valves, see Fig.
Distribution
Holarctic. Known from Canada (Alberta, Northwest territories, British Columbia), Sweden (North Sweden, Abisko,
Ecology
The original description of D. (I.) intricata (
Conservation
Dicranomyia (I.) intricata is red-listed in Finland (NT,
Taxon discussion
See Dicranomyia (I.) boreobaltica Salmela sp.n.
DNA barcode
Standard 5′ region (658 bp) of the cytochrome c oxidase I (COI) gene of Dicranomyia (I.) intricata (BOLD Sample IDs JES-20120082 and JES-20110082, identical specimens):
TACCTTATACTTTATTTTTGGAGCTTGAGCAGGAATAGTAGGAACTTCACTAAGTATTATTATTCGAGCAGAATTAGGACACCCAGGAGCATTAATTGGAGATGACCAAATTTATAATGTAGTAGTTACTGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTCGGTAATTGATTAGTTCCTTTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGAATAAATAATATAAGTTTTTGAATACTTCCCCCTTCTTTAACCTTATTATTAGCTAGAAGTATAGTTGAAAACGGGGCAGGAACTGGTTGAACAGTTTACCCTCCCCTTTCTTCTGGAATTGCTCATTCAGGAGCTTCTGTAGACTTAGCTATTTTTTCTCTTCATTTAGCAGGTATTTCTTCTATTTTAGGAGCTGTTAACTTTATTACAACTGTTATTAATATACGTTCAGCAGGAATTTCATTCGACCGAATACCATTATTTGTTTGATCAGTAGTAATTACTGCTATTCTATTACTCTTATCACTCCCTGTTTTAGCTGGAGCTATTACAATATTATTAACAGATCGAAACTTAAACACTTCATTTTTTGACCCTGCAGGTGGAGGAGATCCTATTTTATACCAACACTTATTT
Analysis
The phylogenetic tree (length 622 steps) resulting from the parsimony analysis is shown in Fig.
Acknowledgements
Barcode sequences were obtained at the Canadian Centre for DNA Barcoding based in the Biodiversity Institute of Ontario at the University of Guelph. Their work was supported by funding from the Government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life Project. Finnish Barcode of Life (FinBOL), led by Dr. Marko Mutanen (Oulu) is also thanked for a help in the barcoding process. JS would like to thank Mr. Teemu Nieminen for his trust and co-operation during his MSc project. Mr. Matti Mäkilä (Rovaniemi) is thanked for his in help in field work and painstaking sorting of Malaise traps samples. Dr. Tapani Sallantaus (Helsinki) kindly gave us his data on water quality from Kusiaiskorpi. KMK was financially supported by Emil Aaltonen foundation. English text was improved by Pete Boardman (Shrewsbury). We also thank referees for their constructive comments.
Author contributions
JS wrote the majority of the manuscript and took some layer photos. VV performed the phylogenetic analyses. KMK took most of the layer photos.
References
- Records and descriptions of crane-flies from Alberta (Tipulidae, Diptera). I.Canadian Entomologist59:214‑225. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- Tipulidae. McAlpine, J.F. et al. (eds).Manual of Nearctic Diptera, Biosystematic Research Centre, Ottawa, Ontario1:153‑190.
- The Phytogeography of Northern Europe.Cambridge University Press,297pp. https://doi.org/10.1017/cbo9780511565182
- Studien über palaearktische, vorwiegend hollandische Limnobiiden, insbesondere über ihre Kopulationsorgane.Tijdschrift voor Entomologie62:52‑97.
- Dicranomyia (Idiopyga) oosterbroeki spec. nov., a new species of Limoniidae (Diptera) from Kazakhstan.Studia Dipterologica20:85‑89. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- deWaard J, Ivanova N, Hajibabaei M, Hebert PN (2008) Assembling DNA Barcodes. In: Martin C (Ed.) Methods in Molecular Biology.Humana Press Inc.,275-293pp. https://doi.org/10.1007/978-1-59745-548-0_15
- PARSIMONY JACKKNIFING OUTPERFORMS NEIGHBOR-JOINING.Cladistics12(2):99‑124. https://doi.org/10.1111/j.1096-0031.1996.tb00196.x
- TNT, a free program for phylogenetic analysis.Cladistics24(5):774‑786. https://doi.org/10.1111/j.1096-0031.2008.00217.x
- Hämet-Ahti L, Suominen J, Ulvinen T, Uotila P (1998) Retkeilykasvio. Field Flora of Finland.3.Kasvimuseo,Helsinki,656pp. [ISBN951-45-8167-9].
- DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions.PLoS ONE6(2):e17134. https://doi.org/10.1371/journal.pone.0017134
- Atlas över växternas utbredning i Norden.Geologiska Föreningan i Stockholm. Foerhandlingar72(3):1‑512. [InSwedish]. https://doi.org/10.1080/11035895009451861
- A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences.Journal of Molecular Evolution16:111‑120. https://doi.org/10.1007/BF01731581
- Species delimitation in the leaf beetle genus Macroplea (Coleoptera, Chrysomelidae) based on mitochondrial DNA, and phylogeographic considerations.Insect Systematics & Evolution37(4):467‑479. https://doi.org/10.1163/187631206788831443
- 1.3. Morphology and terminology of the female postabdomen. in: Papp, L. & Darvas, B. eds. Contributions to a Manual of Palaearctic Diptera (with special reference to flies of economic importance).Science Herald, Budapest1:75‑84.
- 3 Sisävedet ja rannat. In: Raunio, A., Schulman, A. & Kontula, T. (eds.).Suomen luontotyyppien uhanalaisuus – Osa 2: Luontotyyppien kuvaukset2:89‑142. [InFinnish]. URL: https://helda.helsinki.fi/bitstream/handle/10138/37932/SY%208%202008%20Osa%202%203%20Sisavedet%20ja%20rannat.pdf?sequence=4
- Determinants of the biogeographical distribution of butterflies in boreal regions.Journal of Biogeography33(10):1764‑1778. https://doi.org/10.1111/j.1365-2699.2005.01395.x
- Proceedings of the Gateway Computing Environments Workshop (GCE).2010 Gateway Computing Environments Workshop (GCE),8pp. https://doi.org/10.1109/gce.2010.5676129
- Remarks on the life history, female external appearance and conservation status of Elachista vonschantzi Svensson, 1976 (Lepidoptera, Elachistidae) in Fennoscandia.Entomologisk Tidskrift124(3): 183–186.(3):183‑186.
- Wide-ranging barcoding aids discovery of one-third increase of species richness in presumably well-investigated moths.Scientific Reports3:2901. https://doi.org/10.1038/srep02901
- Diagnosen über fünf neue europaische Limoniinae (Dipt. Tipulidae).Zeitschrift der Wiener Entomologischen Gesellschaft38:34‑36.
- Nematoceran flies (Diptera, Nematocera) of the sea-shore meadows and -forests in the northernmost part of the Gulf of Bothnia in Hailuoto and Oulunsalo.MSc thesis, University of Jyväskylä, Finland1:1‑37. [InFinnish, Abstract in English]. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- Graves of Giants and Cairns of Dread: The archaeology of Ostrobothnian stone structures.Acta universitatis ouluensisB52:1‑303. [InFinnish]. URL: http://herkules.oulu.fi/isbn951427170X/isbn951427170X.pdf
- Catalogue of the Craneflies of the World (Diptera, Tipuloidea: Pediciidae, Limoniidae, Cylindrotomidae, Tipulidae).http://nlbif.eti.uva.nl/ccw/index.php2014:8.9.. URL: http://nlbif.eti.uva.nl/ccw/index.php
- Barcoding Bugs: DNA-Based Identification of the True Bugs (Insecta: Hemiptera: Heteroptera).PLoS ONE6(4):e18749. https://doi.org/10.1371/journal.pone.0018749
- Faunistic review of the cuckoo wasps of Fennoscandia, Denmark and the Baltic countries (Hymenoptera: Chrysididae).Zootaxa3864:1‑67.
- Barcoding Beetles: A Regional Survey of 1872 Species Reveals High Identification Success and Unusually Deep Interspecific Divergences.PLoS ONE9(9):e108651. https://doi.org/10.1371/journal.pone.0108651
- Penttinen J, Ilmonen J, Jakovlev J, Salmela J, Kuusela K, Paasivirta L (2010) Thread-horned flies (Diptera: Nematocera). In: Rassi P, Hyvärinen E, Juslen A, Mannerkoski I (Eds) The 2010 Red List of Finnish Species.Ympäristoministeriö & Suomen ympäristökeskus,Helsinki. URL: http://www.ym.fi/fi-FI/Ajankohtaista/Julkaisut/Erillisjulkaisut/Suomen_lajien_uhanalaisuus__Punainen_kir(4709)
- Cryptic animal species are homogeneously distributed among taxa and biogeographical regions.BMC Evolutionary Biology7(1):121. https://doi.org/10.1186/1471-2148-7-121
- Description and DNA barcoding of Tipula (Pterelachisus) recondita sp. n. from the Palaearctic region (Diptera, Tipulidae).ZooKeys192:51. https://doi.org/10.3897/zookeys.192.2364
- New records of crane-flies from NW Russia, with ecological notes on some species (Diptera, Tipulidae, Limoniidae).Zoosystematica Rossica11:361‑366. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- Land uplift phenomenon and its effects on mire vegetation. In: Lindholm, T. & Heikkilä, R. (eds.) Finland - Land of mires.The Finnish Environment23:145‑154. URL: https://helda.helsinki.fi/bitstream/handle/10138/37961/FE_23_2006.pdf?sequence=6
- Biodiversity and community structure of nematoceran flies and mosses of springs in the Lapland triangle region.Master of Science Thesis, University of Jyvaskyla1:1‑56. [InFinnish, English Abstract]. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- Cranefly (Diptera, Tipuloidea & Ptychopteridae) fauna of Limhamn limestone quarry (Sweden, Malmo): Diversity and faunistics viewed from a NW European perspective.Norwegian Journal of Entomology57:123‑135. URL: http://www.entomologi.no/journals/nje/2010-2/pdf/nje-vol57-no2-salmela.pdf
- The semiaquatic nematoceran fly assemblages of tree wetland habitats and concordance with plant species composition, a case study from subalpine Fennoscandia.Journal of Insect Science11 (35):1‑28. URL: http://www.insectscience.org/11.35/
- Annotated list of Finnish crane flies (Diptera: Limoniidae, Tipulidae, Pediciidae & Cylindrotomidae).Entomologica Fennica22:219‑242. URL: http://ojs.tsv.fi/index.php/entomolfennica/article/view/5002/4535
- Biogeographic Patterns of Finnish Crane Flies (Diptera, Tipuloidea).Psyche: A Journal of Entomology2012:1‑20. https://doi.org/10.1155/2012/913710
- Taxonomy, species richness and biogeography of Finnish crane flies (Diptera, Tipuloidea).Annales Universitatis Turkuensis (Series AII)276:1‑41. URL: http://www.doria.fi/handle/10024/87628
- On a stenopterous form of Dicranomyia complicata de Meijere (Diptera, Limoniidae).Dopovidi Akademii Nauk Ukrayinskoyi RSR (B)1970:466‑469. [InUkrainian].
- Idiopyga Savtshenko nom. nov. (Diptera, Limoniidae).Vestnik Zoologii1987(6):81. [InRussian].
- Family Limoniidae.Catalogue of Palaearctic Diptera1:183‑369. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- Perinnebiotoopit. In. Raunio, A. Schulman, A. & Kontula, T. (eds.). Suomen luontotyyppien uhanalaisuus − Osa II: Luontotyyppien kuvaukset., Helsinki.8/2008.Suomen ympäristökeskus,Helsinki,574pp. [InFinnish]. URL: https://helda.helsinki.fi/bitstream/handle/10138/37932/SY%208%202008%20Osa%202%207%20Perinnebiotoopit.pdf?sequence=8
- RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Bioinformatics30(9):1312‑1313. https://doi.org/10.1093/bioinformatics/btu033
- A Rapid Bootstrap Algorithm for the RAxML Web Servers.Systematic Biology57(5):758‑771. https://doi.org/10.1080/10635150802429642
- Revision of European species of the genus Rhabdomastix (Diptera: Limoniidae). Part 2: Subgenus Rhabdomastix s. str.European Journal of Entomology101:657‑687.
- Nomenclatural changes in West Palaearctic Limoniidae and Pediciidae (Diptera), II.Casopis Slezskeho Musea v Opava (A)56:23‑36.
- A new Phyllolabis from Israel with reduced wings and halteres (Diptera: Limoniidae) 41-42: 107-114.Israel Journal of Entomology41-42:107‑114. URL: http://nlbif.eti.uva.nl/ccw/literature.php
- From 'cryptic species' to integrative taxonomy: an iterative process involving DNA sequences, morphology, and behaviour leads to the resurrection of Sepsis pyrrhosoma (Sepsidae: Diptera).Zoologica Scripta39(1):51‑61. https://doi.org/10.1111/j.1463-6409.2009.00408.x
- A synopsis of the Swedish Tipulidae, 1. Subfam. Limoniinae: tribe Limoniini.Opuscula Entomologica23:133‑169.
- Biogeography of Northern European insects: province records in multivariate analysis (Saltatoria; Lepidoptera: Sesiidae; Coleoptera: Buprestidae, Cerambycidae). 28: 57–81.Annales Zoologi Fennici28:57‑81.
- Integrative taxonomy, or iterative taxonomy?Systematic Entomology36(2):209‑217. https://doi.org/10.1111/j.1365-3113.2010.00558.x
Supplementary materials
Non-type material of Dicranomyia (I.) intricata Alexander, males, permanently slide-mounted by C.P. Alexander, deposited in USNM (USA, Washington). Digital photos of the slides.
COI 5' standard DNA barcoding fragment