|
Biodiversity Data Journal :
Software Description
|
|
Corresponding author: Edward Baker (edwbaker@gmail.com)
Academic editor: Anne Thessen
Received: 03 Oct 2019 | Accepted: 21 Nov 2019 | Published: 27 Nov 2019
© 2019 Edward Baker, Steen Dupont, Vincent Smith
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Baker E, Dupont S, Smith VS (2019) Ecological interactions in the Scratchpads virtual research environment. Biodiversity Data Journal 7: e47043. https://doi.org/10.3897/BDJ.7.e47043
|
|
Abstract
Background
The Natural History Museum, London has a number of online databases that describe interactions between species, including the HOSTS database of lepidopteran host plants (
New information
This paper describes the implementation within the Scratchpads VRE of a new ecological interactions module that is capable of handling the needs of these projects, while at the same time is flexible to handle the needs of future projects with different data sources.
Keywords
ecological informatics, biodiversity informatics, ecological interactions
Introduction
In order to understand life on Earth, it is essential to understand not only the distribution and traits of species, but how they interact with each other. Biodiversity informatics as a discipline has created global infrastructures for both species distribution (GBIF) and trait (TraitBank) data. Global Biotic Interactions (GloBI:
Ecological interaction datasets are, like natural history specimens, fragmented and widely distributed. They can be found scattered through scientific literature and specimen labels in museums. Numerous publications have synthesised interaction datasets for taxonomic groups (e.g. cockroaches
The need for biodiversity informatics to address species interactions was set as a challenge by
Project description
The existing datasets to be migrated were all of a similar format: two species had been documented interacting in a bibliographic reference, often at a specified location. While this model formed the basis of the implementation, we expanded this model to allow for interactions that are recorded from museum specimens (for an example see
The introduction of non-native species into an ecosystem may result in novel interactions behaviour. For example, the accidental introduction of the stick insect Carausius morosus (Sinéty, 1901) into the San Diego area means it has been found on many plants it would not have encountered in its native habitat in southern India (
Organism status (presence status; aligned to DarwinCore establishmentMeans).
| Status | Description |
|---|---|
| Native | The organism either evolved in this region or arrived by non-anthropogenic means. |
| Naturalised | The organism reproduces naturally and forms part of the local ecology. |
| Introduced | The organism arrived in the region via an anthropogenic mechanism or mechanisms. |
| Invasive | The organism is having a deleterious impact on another organism, multiple organisms or the ecosystem as a whole. |
| Captivity | The organism is kept in captivity. |
| Managed | The organism maintains its presence through intentional cultivation or husbandry. |
Another example of potentially confusing data for phasmids is the numerous food plants that are successfully used to rear these species in captivity (e.g.
| Status | Description |
| Interaction recorded in the wild | |
| Interaction recorded in captivity | Used when the status of the specimens is uncertain |
| Interaction recorded in captivity from wild caught specimens | |
| Interaction recorded in captivity from captive bred specimens |
In addition, some interactions have significant importance, such as the defoliation of food and timber crops by stick insects (
| Status | Description |
| Economic | The interaction has financial impact for human society |
| Economic (crop pest) | The interaction is damaging to food crops |
| Economic (timber pest) | The interaction is damaging to timber |
| Economic (pest control) | The interaction helps to control a pest species |
| Medical | The interaction has medical important consequences on humans |
| Veterinary | The interaction has medical important consequences on animals |
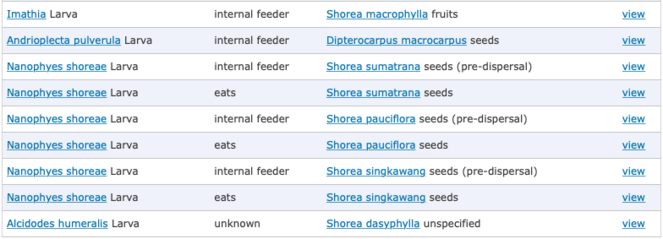
The migrated hosts data (
The above examples are limited to the Phasmida, a small order of insects with less than 4,000 valid species (
The Phthiraptera Scratchpad (http://phthiraptera.info/) documents approximately 12,000 interactions between parasitic lice (Subinfraorder Phthiraptera) and their mammal and bird hosts. Multiple mammal classifications are used, reflecting the fact that different authorities have used different host classifications when compiling checklists for blood sucking lice (superfamily Anoplura) and chewing lice (superfamilies Amblycera, Ischnocera and Rhynchophthirina). This extensive database underpins a significant body of research on parasitic lice, which is used as a model to study co-speciation.
The Scratchpads project was conceived and developed as part of a much wider portfolio of biodiversity informatics platforms and systems, so from the outset, the system described here was designed to operate with the Global Biotic Interactions project (GloBI;
Funding was provided by the Natural History Museum to employ EB during part of 2016.
Web location (URIs)
Technical specification
Repository
Usage rights
The code developed for this project is, like the rest of the Scratchpads project, released under the GNU General Public License v2.0. Scratchpad users have fine-grain control over the licence applied to each piece of content on their site, but the project encourages the use of open data licences following
Additional information
Implementation
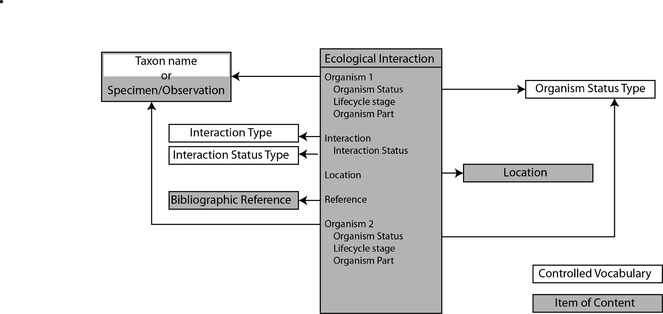
The implementation creates a new content type (Drupal: node type) for ecological interactions. Each interaction (a Drupal node) has a unique URL and identifier (UUID). The UUID uniquely identifies each interaction and should persist if the dataset is aggregated, as a means to trace the provenance of the data in the source dataset (e.g. to correct errors or add annotation). The Scratchpads enivronment has defined content types for bibliographic references, specimens and observations and locations, as well as tools for manipulating biological classification. The new ecological interactions content type links to these existing content types and classifications as shown in Fig.
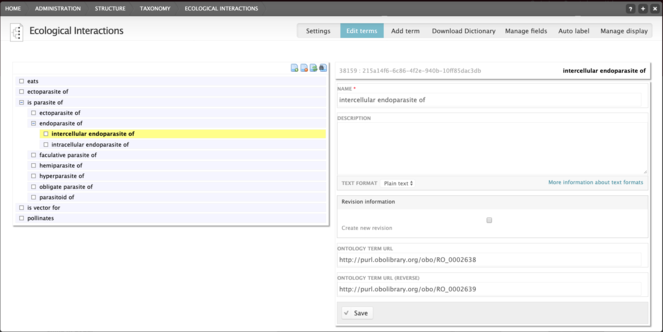
The Relations Ontology (RO; https://www.ebi.ac.uk/ols/ontologies/ro) provides several ontology terms for describing inter-species interactions, which have been adopted by the module described here. The system presents a human-readable description (e.g. "eats") to the user, but additionally stores the relevant URI from the RO. In addition, the reverse URI is also stored (e.g. "eaten by" is the reverse of "eat"). At present, this functionality is not used, but in the future, this will allow for more advanced searches. These terms are stored in a Drupal taxonomy, allowing terms to be stored in a hierarchy (i.e. "ectoparasite of" is a child term of "parasite of"; Fig.
To maintain visual coherence with other Scratchpads features and for increased search speed, the main ecological interactions page (showing all interactions; Fig.
A text file suitable for ingest by GloBI is found at /interactions.txt on Scratchpad sites where the module is enabled (e.g. http://phthiraptera.info/interactions.txt). This file can be harvested by external aggregators.
Future plans
The NHM is undertaking work to combine the output of several Scratchpad sites, as well as other sources, to create an institutional 'interactions bank' that will provide a unified entry point for these disparate interactions datasets. The NHM Data Portal is used by NHM staff to publish their research datasets, whereas Scratchpads can be used by both NHM and external researchers. For this reason, it is currently possible to contribute to GloBI directly from an individual Scratchpad and NHM-affiliated researchers, in future, will be able to contribute via the interactions bank.
An example of a Scratchpad hosted project, that is starting to adopt the ecological interactions module, is the BioAcoustica database (
The interactions of species with the human environment is also not yet properly covered.
The data model we have developed is based upon the needs of the initial projects migrated and there is scope for future additions to accommodate additional needs (e.g. recording the date and time of observed interactions using DarwinCore eventTime).
There is great potential for the biodiversity informatics community to adopt or develop controlled vocabularies.
Acknowledgements
Jorrit Poelen has provided advice at all stages of the project. In addition, we would like to thank the authors and compilers of the projects that have been migrated.
Author contributions
The project to move existing databases into modern infrastructures was overseen by VS. EB researched the requirements and implemented the Scratchpad module. SD has oversight of the HOSTS project and advised on requirements during development.
References
- The Food Plants of Phasmids.Practical Reptile Keeping(13)44‑46.
- Carausius morosus in San Diego.Figsharehttps://doi.org/10.6084/M9.FIGSHARE.1304202.V1
- The worldwide status of phasmids (Insecta: Phasmida) as pests of agriculture and forestry, with a generalised theory of phasmid outbreaks.Agriculture & Food Security4(1). https://doi.org/10.1186/s40066-015-0040-6
- BioAcoustica: a free and open repository and analysis platform for bioacoustics.Database2015https://doi.org/10.1093/database/bav054
- Controlled values for Establishment Means from Darwin Core. http://baskauf.blogspot.com/2016/04/controlled-values-for-establishment.html. Accessed on: 2019-11-07.
- Variation of butterfly diet breadth in relation to host-plant predictability: results from two faunas.Oikos90(1):50‑66. https://doi.org/10.1034/j.1600-0706.2000.900106.x
- Catalogue of the Hostplants of the Neotropical Butterflies. [Catálogo de las Plantas Huésped de las Mariposas Neotropicales].Sociedad Entomológica Aragonesa (SEA),Zaragoza, Spain. [ISBN978-84-935872-2-2]
- Food plant selection by stick insects (Phasmida) in a Bornean rain forest.Journal of Tropical Ecology22(01):35‑40. https://doi.org/10.1017/s0266467405002749
- Caterpillars and Host Plant Records for 59 Species of Geometridae (Lepidoptera) from a Montane Rainforest in Southern Ecuador.Journal of Insect Science10(67):1‑22. https://doi.org/10.1673/031.010.6701
- Phasmida SpeciesFile Online. http://phasmida.speciesfile.org. Accessed on: 2016-6-10.
- Patterns of Lepidoptera herbivory on conifers in the New World.Journal of Asia-Pacific Biodiversity11(1):1‑10. https://doi.org/10.1016/j.japb.2018.01.008
- A hymenopterists’ guide to the Hymenoptera Anatomy Ontology: utility, clarification, and future directions.Journal of Hymenoptera Research27:67‑88. https://doi.org/10.3897/jhr.27.2961
- NHM Interactions Bank.Figsharehttps://doi.org/10.6084/M9.FIGSHARE.7252583.V1
- Creative Commons licenses and the non-commercial condition: Implications for the re-use of biodiversity information.ZooKeys150:127‑149. https://doi.org/10.3897/zookeys.150.2189
- A decadal view of biodiversity informatics: challenges and priorities.BMC Ecology13(1). https://doi.org/10.1186/1472-6785-13-16
- Repeated adaptive divergence of microhabitat specialization in avian feather lice.BMC Biology10(1). https://doi.org/10.1186/1741-7007-10-52
- Feeding Preferences of Phasmids (Insecta: Phasmida) in a Bornean Dipterocarp Forest.The Raffles Bulletin of Zoology56:445‑452.
- Developing a dedicated cestode life cycle database: lessons from the hymenolepidids.Helminthologia46(1):21‑27. https://doi.org/10.2478/s11687-009-0004-0
- Uberon, an integrative multi-species anatomy ontology.Genome Biology13(1). https://doi.org/10.1186/gb-2012-13-1-r5
- Global biotic interactions: An open infrastructure to share and analyze species-interaction datasets.Ecological Informatics24:148‑159. https://doi.org/10.1016/j.ecoinf.2014.08.005
- The Suitability of Select Ferns as Hosts for Archips machlopis (Lepidoptera: Tortricidae).Florida Entomologist99(3):572‑573. https://doi.org/10.1653/024.099.0341
- Acoustic attraction of the parasitoid fly Ormia ochracea to the song of its host.The Journal of the Acoustical Society of America105(2):1319‑1320. https://doi.org/10.1121/1.424543
- HOSTS - A Database of the World's Lepidopteran Hostplants. Natural History Museum, London.https://www.nhm.ac.uk/our-science/data/hostplants/. Accessed on: 2016-8-20.
- The Biotic Associations of Cockroaches.Smithsonian Miscellaneous Collections141.
- The Natural History Museum Data Portal.Database2019https://doi.org/10.1093/database/baz038
- phthiraptera.info. http://phthiraptera.info. Accessed on: 2019-5-29.
- Scratchpads 2.0: a Virtual Research Environment supporting scholarly collaboration, communication and data publication in biodiversity science.ZooKeyshttps://doi.org/10.3897/zookeys.150.2193
- Evolution of Gustatory Receptor Gene Family Provides Insights into Adaptation to Diverse Host Plants in Nymphalid Butterflies.Genome Biology and Evolution10(6):1351‑1362. https://doi.org/10.1093/gbe/evy093
- Phylogenetic indices for measuring the diet breadths of phytophagous insects.Oecologia119(3):427‑434. https://doi.org/10.1007/s004420050804
- TaxonWorks: A Use Case in Documenting Complex Biological Relationships.Biodiversity Information Science and Standards2https://doi.org/10.3897/biss.2.25723