|
Biodiversity Data Journal :
Taxonomic paper
|
|
Corresponding author:
Academic editor: Pavel Stoev
Received: 24 Aug 2015 | Accepted: 24 Aug 2015 | Published: 30 Aug 2015
© 2015 Angela Telfer, Monica Young, Jenna Quinn, Kate Perez, Crystal Sobel, Jayme Sones, Valerie Levesque-Beaudin, Rachael Derbyshire, Jose Fernandez-Triana, Rodolphe Rougerie, Abinah Thevanayagam, Adrian Boskovic, Alex Borisenko, Alex Cadel, Allison Brown, Anais Pages, Anibal Castillo, Annegret Nicolai, Barb Mockford Glenn Mockford, Belén Bukowski, Bill Wilson, Brock Trojahn, Carole Ann Lacroix, Chris Brimblecombe, Christoper Hay, Christmas Ho, Claudia Steinke, Connor Warne, Cristina Garrido Cortes, Daniel Engelking, Danielle Wright, Dario Lijtmaer, David Gascoigne, David Hernandez Martich, Derek Morningstar, Dirk Neumann, Dirk Steinke, Donna DeBruin Marco DeBruin, Dylan Dobias, Elizabeth Sears, Ellen Richard, Emily Damstra, Evgeny Zakharov, Frederic Laberge, Gemma Collins, Gergin Blagoev, Gerrie Grainge, Graham Ansell, Greg Meredith, Ian Hogg, Jaclyn McKeown, Janet Topan, Jason Bracey, Jerry Guenther, Jesse Sills-Gilligan, Joseph Addesi, Joshua Persi, Kara Layton, Kareina D'Souza, Kencho Dorji, Kevin Grundy, Kirsti Nghidinwa, Kylee Ronnenberg, Kyung Min Lee, Linxi Xie, Liuqiong Lu, Lyubomir Penev, Mailyn Gonzalez, Margaret Rosati, Mari Kekkonen, Maria Kuzmina, Marianne Iskandar, Marko Mutanen, Maryam Fatahi, Mikko Pentinsaari, Miriam Bauman, Nadya Nikolova, Natalia Ivanova, Nathaniel Jones, Nimalka Weerasuriya, Norman Monkhouse, Pablo Lavinia, Paul Jannetta, Priscila Hanisch, R. Troy McMullin, Rafael Ojeda Flores, Raphaëlle Mouttet, Reid Vender, Renee Labbee, Robert Forsyth, Rob Lauder, Ross Dickson, Ruth Kroft, Scott Miller, Shannon MacDonald, Sishir Panthi, Stephanie Pedersen, Stephanie Sobek-Swant, Suresh Naik, Tatsiana Lipinskaya, Thanushi Eagalle, Thibaud Decaëns, Thibault Kosuth, Thomas Braukmann, Tom Woodcock, Tomas Roslin, Tony Zammit, Victoria Campbell, Vlad Dinca, Vlada Peneva, Paul Hebert, Jeremy deWaard
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Telfer A, Young M, Quinn J, Perez K, Sobel C, Sones J, Levesque-Beaudin V, Derbyshire R, Fernandez-Triana J, Rougerie R, Thevanayagam A, Boskovic A, Borisenko A, Cadel A, Brown A, Pages A, Castillo A, Nicolai A, Glenn Mockford B, Bukowski B, Wilson B, Trojahn B, Lacroix C, Brimblecombe C, Hay C, Ho C, Steinke C, Warne C, Garrido Cortes C, Engelking D, Wright D, Lijtmaer D, Gascoigne D, Hernandez Martich D, Morningstar D, Neumann D, Steinke D, Marco DeBruin D, Dobias D, Sears E, Richard E, Damstra E, Zakharov E, Laberge F, Collins G, Blagoev G, Grainge G, Ansell G, Meredith G, Hogg I, McKeown J, Topan J, Bracey J, Guenther J, Sills-Gilligan J, Addesi J, Persi J, Layton K, D'Souza K, Dorji K, Grundy K, Nghidinwa K, Ronnenberg K, Lee K, Xie L, Lu L, Penev L, Gonzalez M, Rosati M, Kekkonen M, Kuzmina M, Iskandar M, Mutanen M, Fatahi M, Pentinsaari M, Bauman M, Nikolova N, Ivanova N, Jones N, Weerasuriya N, Monkhouse N, Lavinia P, Jannetta P, Hanisch P, McMullin R, Ojeda Flores R, Mouttet R, Vender R, Labbee R, Forsyth R, Lauder R, Dickson R, Kroft R, Miller S, MacDonald S, Panthi S, Pedersen S, Sobek-Swant S, Naik S, Lipinskaya T, Eagalle T, Decaëns T, Kosuth T, Braukmann T, Woodcock T, Roslin T, Zammit T, Campbell V, Dinca V, Peneva V, Hebert P, deWaard J (2015) Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve. Biodiversity Data Journal 3: e6313. https://doi.org/10.3897/BDJ.3.e6313
|
|
Abstract
Background
Comprehensive biotic surveys, or ‘all taxon biodiversity inventories’ (ATBI), have traditionally been limited in scale or scope due to the complications surrounding specimen sorting and species identification. To circumvent these issues, several ATBI projects have successfully integrated DNA barcoding into their identification procedures and witnessed acceleration in their surveys and subsequent increase in project scope and scale. The Biodiversity Institute of Ontario partnered with the rare Charitable Research Reserve and delegates of the 6th International Barcode of Life Conference to complete its own rapid, barcode-assisted ATBI of an established land trust in Cambridge, Ontario, Canada.
New information
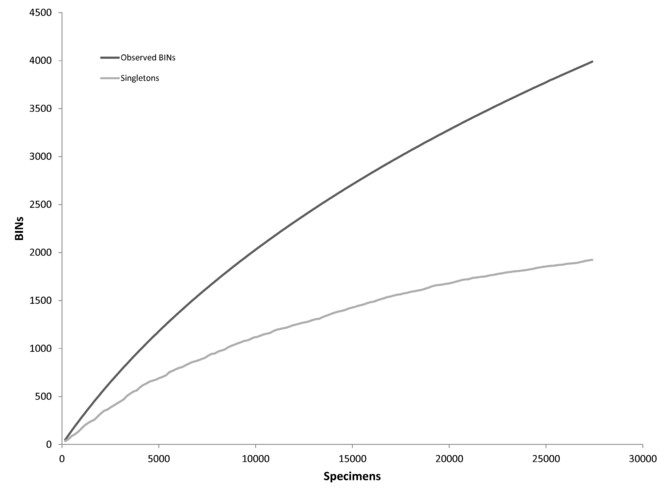
The existing species inventory for the rare Charitable Research Reserve was rapidly expanded by integrating a DNA barcoding workflow with two surveying strategies – a comprehensive sampling scheme over four months, followed by a one-day bioblitz involving international taxonomic experts. The two surveys resulted in 25,287 and 3,502 specimens barcoded, respectively, as well as 127 human observations. This barcoded material, all vouchered at the Biodiversity Institute of Ontario collection, covers 14 phyla, 29 classes, 117 orders, and 531 families of animals, plants, fungi, and lichens. Overall, the ATBI documented 1,102 new species records for the nature reserve, expanding the existing long-term inventory by 49%. In addition, 2,793 distinct Barcode Index Numbers (BINs) were assigned to genus or higher level taxonomy, and represent additional species that will be added once their taxonomy is resolved. For the 3,502 specimens, the collection, sequence analysis, taxonomic assignment, data release and manuscript submission by 100+ co-authors all occurred in less than one week. This demonstrates the speed at which barcode-assisted inventories can be completed and the utility that barcoding provides in minimizing and guiding valuable taxonomic specialist time. The final product is more than a comprehensive biotic inventory – it is also a rich dataset of fine-scale occurrence and sequence data, all archived and cross-linked in the major biodiversity data repositories. This model of rapid generation and dissemination of essential biodiversity data could be followed to conduct regional assessments of biodiversity status and change, and potentially be employed for evaluating progress towards the Aichi Targets of the Strategic Plan for Biodiversity 2011–2020.
Keywords
DNA barcoding, species identification, biodiversity assessment, biotic inventory, Barcode Index Numbers, Operational Taxonomic Units, rare Charitable Research Reserve
Introduction
It is now universally accepted that we have entered a period of unprecedented global biodiversity loss (
Even prior to the concept’s introduction (
Following this model, the present study introduces DNA barcoding to a long-term biotic inventorying effort being conducted in a temperate nature reserve. The objective is to gauge the effect of adding this tool, in terms of both acceleration and increase of taxonomic scope, while concurrently constructing a reference DNA barcode library to facilitate future research and monitoring at this site. The existing inventory is expanded by employing two surveying strategies – a longer and comprehensive invertebrate trapping scheme, followed by a concentrated effort involving taxonomic experts in the form of a bioblitz (
Materials and methods
Study Site and Existing Species Inventory
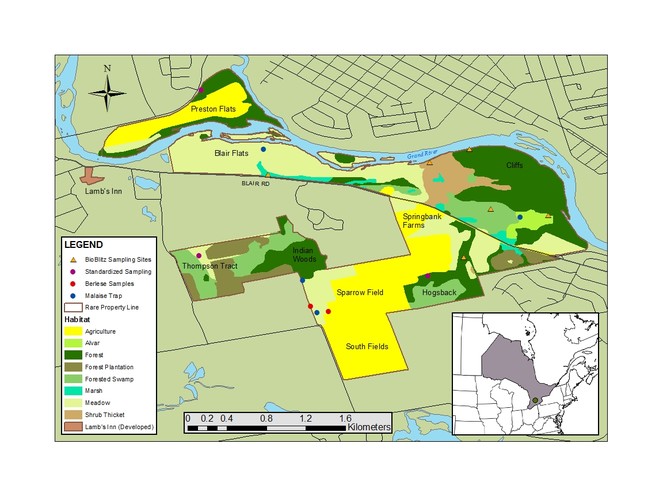
The rare Charitable Research Reserve is a 365+ hectare land reserve which was set aside in 2001 to preserve the cultural history and ecological integrity of the area, while providing opportunities for scientific research and public education within the context of an urbanized region. It is located at the confluence of the Speed and Grand Rivers in Cambridge, Ontario, Canada (43.381128, -80.357807), where the Carolinian and Northern Hardwood forests also meet. The reserve contains a diversity of habitats including existing and reclaimed agricultural lands, wetlands, floodplains, shrub thickets, limestone cliffs and alvars, cold-water creeks, and old growth forest. Due to these diverse habitats, as well as the organization’s mandate to facilitate scientific research, rare has been the site of a variety of innovative research studies, including studies on fern genetics (
Prior to this study, 2,246 species had been recorded at rare, including birds (231), mammals (37), insects (832), plants (836), mosses (63) and lichens (21) (Suppl. material
Survey Strategies and Specimen Collection
Two strategies were employed in an effort to maximize the diversity of organisms inventoried. The first was a comprehensive collecting scheme executed over a period of approximately four months (May to August 2015). It involved a variety of targeted taxa and techniques, but heavily favoured the collection of terrestrial arthropods by passive trapping. Four Malaise traps were set up in various habitats around the rare property (Fig.
The second strategy for surveying the reserve was a more concentrated effort and involved taxonomic experts – the execution of a bioblitz (
DNA Barcode Analysis
Both surveying strategies provided a large number of specimens that were sorted and prepared for subsequent DNA barcode analyses at the Canadian Centre for DNA barcoding (CCDB; www.ccdb.ca). A total of 25,287 specimens were sequenced from collection efforts from May to August 2015, followed by 3,502 specimens directly following the rare BioBlitz on Aug 16, 2015. Tissue samples were prepared in 96-well plate format and when necessary, the whole specimen proceeded through lysis and was recovered as voucher from the filter plate (
One or more standard DNA barcode markers were targeted for each major group of organisms: for animals, the mitochondrial gene cytochrome oxidase subunit 1 (COI) (
Barcode Index Numbers and Taxonomic Assignment
For the sequences derived from animal specimens, the records were assigned operational taxonomic units (OTUs) called Barcode Index Numbers (BINs) by the Refined Single Linkage (RESL) algorithm implemented on BOLD (
For each specimen that was assigned an existing BIN, the record received the existing identification of the BIN to the lowest level that did not have taxonomic conflict. For each specimen assigned a new BIN for BOLD, the sequence was queried through the BOLD Identification Engine (BOLD-ID Engine; http://www.boldsystems.org/index.php/IDS_OpenIdEngine). Identifications were applied based on sequence similarity (<15% for family, <5% for genus) if the query sequence fell within a monophyletic cluster of BINs assigned to this family or genus. For animal records that did not receive BINs (<500bp), the sequence was similarly queried through the BOLD-ID Engine, but used a <2% similarity cutoff for assignment to species, in addition to the genus and family thresholds. Following this, a neighbour-joining tree was constructed and examined for unexpected placements which might indicate overlooked contamination events or analytical error. Finally, specimens and images were inspected morphologically to check for errors and refine the assigned taxonomy where possible.
Data resources
Collection data, taxonomic assignment, sequence, electropherograms and primer details for each specimen record, and often a high resolution image, are available on BOLD in the public dataset, "rare BioBlitz 2015 [DS-RBB15]" (https://doi.org/10.5883/DS-RBB15 or http://boldsystems.org/index.php/MAS_Management_OpenDataSet?datasetcode=DS-RBB15). The sequence data for each successfully barcoded specimen were deposited to GenBank by using the 'Submit to GenBank' function in the BOLD workbench (see Suppl. material
With the 'Data Spreadsheets' function in the BOLD workbench, the complete dataset was downloaded and reformatted into a Darwin Core Archive (Suppl. material
Telfer A, Young MR, Quinn J, Perez K, Sobel CN, Sones JE, Levesque-Beaudin V, Derbyshire R, Fernandez-Triana J, Rougerie R, Hebert PDN, deWaard JR and contributors* (2015+). Inventory and BioBlitz Records from rare Charitable Research Reserve. 28,916 records. Online at http://data.canadensys.net/ipt/resource.do?r=rare_inventory, http://doi.org/10.5886/hh6td9jn, and http://www.gbif.org/dataset/09e90dfb-5b1b-4dd9-a796-e2fba53d26f0, released on 2015-08-20, version 1. GBIF key: 09e90dfb-5b1b-4dd9-a796-e2fba53d26f0.
* See Suppl. material
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The first of five checklists for Kingdom Animalia, this checklist contains members of Phylum Annelida and Phylum Arthopoda (Class Arachnida and Class Insecta up to Order Dermaptera).
Kingdom
Phylum
Class
Order
Family
Erpobdella punctata
Order
Family
Lumbricus terrestris
Phylum
Class
Order
Family
Agelenopsis potteri
Family
Callobius bennetti
Family
Anyphaena celer
Anyphaena pectorosa
Hibana gracilis
Wulfila saltabundus
Family
Acanthepeira stellata
Araneus diadematus
Araneus trifolium
Araniella displicata
Argiope aurantia
Argiope trifasciata
Eustala anastera
Eustala cepina
Eustala emertoni
Hypsosinga pygmaea
Hypsosinga rubens
Larinioides cornutus
Larinioides patagiatus
Mangora gibberosa
Mangora maculata
Mangora placida
Neoscona arabesca
Family
Clubiona abboti
Clubiona bryantae
Clubiona johnsoni
Clubiona maritima
Clubiona obesa
Clubiona pallidula
Family
Cicurina brevis
Cicurina itasca
Cicurina pallida
Dictyna bellans
Dictyna bostoniensis
Dictyna brevitarsa
Dictyna foliacea
Dictyna volucripes
Emblyna annulipes
Emblyna hentzi
Emblyna manitoba
Emblyna sublata
Family
Drassyllus depressus
Drassyllus niger
Gnaphosa parvula
Haplodrassus signifer
Herpyllus ecclesiasticus
Micaria pulicaria
Sergiolus ocellatus
Zelotes hentzi
Zelotes pseustes
Family
Neoantistea agilis
Neoantistea gosiuta
Family
Agyneta fabra
Agyneta micaria
Agyneta unimaculata
Bathyphantes brevis
Bathyphantes pallidus
Centromerus sylvaticus
Ceraticelus atriceps
Ceraticelus similis
Ceratinella brunnea
Ceratinops crenatus
Ceratinops latus
Ceratinopsis auriculata
Ceratinopsis labradorensis
Collinsia plumosa
Eridantes erigonoides
Erigone atra
Erigone autumnalis
Erigone blaesa
Frontinella communis
Grammonota angusta
Grammonota inornata
Hypomma marxi
Hypselistes florens
Mermessus index
Mermessus trilobatus
Microlinyphia mandibulata
Neriene clathrata
Neriene montana
Neriene variabilis
Pityohyphantes costatus
Pityohyphantes subarcticus
Pocadicnemis americana
Tennesseellum formica
Tenuiphantes zebra
Walckenaeria directa
Walckenaeria fallax
Walckenaeria pinocchio
Walckenaeria spiralis
Family
Arctosa emertoni
Pardosa distincta
Pardosa milvina
Pardosa modica
Pardosa moesta
Pardosa saxatilis
Pirata piraticus
Pirata praedo
Piratula cantralli
Piratula minuta
Schizocosa avida
Schizocosa crassipalpata
Schizocosa mccooki
Schizocosa ocreata
Trochosa ruricola
Trochosa terricola
Family
Mimetus epeiroides
Mimetus haynesi
Mimetus notius
Family
Philodromus cespitum
Philodromus imbecillus
Philodromus praelustris
Philodromus rufus subsp. vibrans
Thanatus coloradensis
Tibellus maritimus
Tibellus oblongus
Family
Phrurotimpus borealis
Scotinella pugnata
Family
Dolomedes striatus
Dolomedes tenebrosus
Pisaurina mira
Family
Eris militaris
Evarcha hoyi
Habronattus decorus
Marpissa formosa
Naphrys pulex
Neon nelli
Pelegrina galathea
Pelegrina insignis
Pelegrina proterva
Phidippus audax
Phidippus clarus
Salticus scenicus
Sitticus floricola subsp. palustris
Synageles noxiosus
Tutelina harti
Tutelina similis
Zygoballus nervosus
Family
Leucauge venusta
Pachygnatha dorothea
Pachygnatha tristriata
Pachygnatha xanthostoma
Tetragnatha caudata
Tetragnatha extensa
Tetragnatha guatemalensis
Tetragnatha laboriosa
Tetragnatha shoshone
Tetragnatha straminea
Tetragnatha viridis
Family
Dipoena nigra
Enoplognatha caricis
Enoplognatha ovata
Euryopis funebris
Hentziectypus globosus
Neospintharus trigonum
Neottiura bimaculata
Parasteatoda tabulata
Parasteatoda tepidariorum
Theridion albidum
Theridion differens
Theridion glaucescens
Theridion murarium
Theridula emertoni
Thymoites unimaculatus
Yunohamella lyrica
Family
Mecaphesa asperata
Misumena vatia
Misumenoides formosipes
Misumessus oblongus
Ozyptila americana
Ozyptila praticola
Tmarus angulatus
Xysticus bicuspis
Xysticus discursans
Xysticus elegans
Xysticus emertoni
Xysticus funestus
Xysticus luctans
Xysticus punctatus
Xysticus winnipegensis
Family
Uloborus glomosus
Order
Family
Arctoseius cetratus
Family
Rhodacarellus silesiacus
Order
Family
Oligolophus tridens
Phalangium opilio
Platybunus triangularis
Family
Leiobunum aldrichi
Leiobunum ventricosum
Order
Family
Euzetes globulus
Family
Gustavia microcephala
Family
Dorycranosus acutidens
Family
Podoribates pratensis
Family
Punctoribates punctum
Family
Oppia nitens
Family
Oribatula tibialis
Family
Scheloribates clavilanceolatus
Family
Tectocepheus sarekensis
Order
Family
Arrenurus reflexus
Class
Order
Family
Bosmina liederi
Family
Eurycercus longirostris
Class
Order
Family
Schendyla nemorensis
Order
Family
Lithobius microps
Class
Entomobrya atrocincta
Family
Entomobrya nivalis
Lepidocyrtus paradoxus
Orchesella villosa
Pseudosinella octopunctata
Family
Parisotoma notabilis
Order
Family
Ceratophysella bengtssoni
Order
Family
Dicyrtomina minuta
Family
Sminthurinus elegans
Brachyiulus pusillus
Family
Cylindroiulus caeruleocinctus
Julus scandinavius
Ophyiulus pilosus
Class
Order
Family
Caenocara sp.
Ptilinus ruficornis
Family
Malporus formicarius
Notoxus desertus
Stricticomus tobias
Family
Anthribus nebulosus
Ormiscus walshii
Family
Agrilus sulciollis
Family
Simplocaria semistriata
Family
Byturus unicolor
Family
Cantharis rufa
Chauliognathus pensylvanicus
Rhagonycha fulva
Rhaxonycha carolina
Family
Agonoleptus conjunctus
Agonum fidele
Agonum gratiosum
Amara angustata
Amara rubrica
Bembidion frontale
Bembidion obtusum
Calleida punctata
Carabus granulatus
Carabus nemoralis
Chlaenius tricolor
Clivina fossor
Colliuris pensylvanica
Dyschirius setosus
Elaphrus clairvillei
Lebia fuscata
Lebia solea
Lebia viridis
Platynus hypolithos
Pterostichus melanarius
Family
Acalymma vittata
Astylopsis macula
Astylopsis sexguttata
Bellamira scalaris
Calligrapha californica subsp. coreopsivora
Chaetocnema sp.
Charidotella sexpunctata subsp. bicolor
Chrysochus auratus
Chrysolina hudsonica
Cicindela punctulata subsp. punctulata
Cicindela sexguttata
Clytus ruricola
Crepidodera sp.
Cyrtophorus verrucosus
Deloyala guttata
Dibolia sp.
Epitrix sp.
Euderces picipes
Gaurotes cyanipennis
Longitarsus sp.
Mantura floridana
Megacyllene robiniae
Molorchus bimaculatus
Neoclytus acuminatus subsp. acuminatus
Oberea tripunctata
Obrium rufulum
Oulema palustris
Paria sp.
Phyllotreta sp.
Phymatodes amoenus
Plagiodera versicolor
Psylliodes punctulata
Pyrrhalta sp.
Systena blanda
Tetraopes tetrophthalmus
Tetrops praeusta
Trigonarthris proxima
Trirhabda sp.
Typocerus velutinus subsp. velutinus
Urgleptes querci
Xylotrechus convergens
Xylotrechus gemellus
Zeugophora varians
Family
Philothermus glabriculus
Family
Altica chalybea
Altica sp.
Cassida rubiginosa
Crepidodera heikertingeri
Dibolia borealis
Epitrix cucumeris
Exema canadensis
Helocassis clavata
Microrhopala vittata
Neogalerucella calmariensis
Ophraella conferta
Paria fragariae
Phyllotreta striolata
Plagiodera versicolora
Psylliodes affinis
Psylliodes napi
Psylliodes picinus
Pyrrhalta viburni
Trirhabda borealis
Xanthonia decemnotata
Family
Ceracis thoracicornis
Family
Cymatodera bicolor
Enoclerus nigripes
Enoclerus rosmarus
Isohydnocera curtipennis
Phyllobaenus verticalis
Placopterus thoracicus
Zenodosus sanguineus
Family
Coccinella septempunctata
Coleomegilla maculata
Coleomegilla maculata subsp. lengi
Didion punctatum
Harmonia axyridis
Hippodamia glacialis
Hippodamia variegata
Hyperaspis binotata
Hyperaspis cf. binotata
Propylaea quatuordecimpunctata
Propylea quatuordecimpunctata
Psyllobora vigintimaculata subsp. maculata
Scymnus sp.
Stethorus punctillum
Family
Orthoperus scutellaris
Sericoderus lateralis
Family
Atomaria ephippiata
Family
Acoptus suturalis
Barypeithes pellucidus
Hylesinus aculeatus
Hypera zoilus
Isochnus sequensi
Madarellus undulatus
Monarthrum mali
Orchestes alni
Phyllobius oblongus
Pityogenes hopkinsi
Polydrusus impressifrons
Sitona lineellus
Tychius meliloti
Xyleborinus alni
Xyleborinus saxeseni
Xyleborus dispar
Xylosandrus germanus
Family
Desmopachria convexa
Family
Aeolus mellillus
Ampedus areolatus
Ampedus linteus
Ampedus nigricollis
Ampedus oblessus
Ampedus protervus
Athous brightwelli
Corymbitodes tarsalis
Ctenicera cylindriformis
Dalopius vagus
Hemicrepidius memnonius
Melanotus castanipes
Family
Mycetina perpulchra
Phymaphora pulchella
Family
Triplax flavicollis
Triplax thoracica
Tritoma pulchra
Tritoma sanguinipennis
Family
Dirrhagofarsus lewisi
Hylis terminalis
Isorhipis obliqua
Microrhagus sp.
Microrhagus subsinuata
Microrhagus triangularis
Family
Dineutus assimilis
Family
Haliplus immaculicollis
Family
Anacaena lutescens
Cercyon haemorrhoidalis
Enochrus ochraceus
Tropisternus natator
Family
Brachypterolus pulicarius
Family
Ellychnia corrusca
Lucidota atra
Photinus sp.
Pyractomena sp.
Pyropyga nigricans
Family
Acropteroxys gracilis
Family
Cortinicara gibbosa
Family
Anisotoma obsoleta
Anisotoma sp.
Catops paramericanus
Catops sp.
Lionothus sp.
Nemadus sp.
Prionochaeta opaca
Ptomophagus sp.
Sciodrepoides fumatus subsp. terminans
Family
Dircaea liturata
Epicauta pensylvanica
Melandrya striata
Family
Epicauta murina
Meloe impressus
Family
Collops quadrimaculata
Hypebaeus apicalis
Family
Mordellina infima
Mordellistena andreae
Mordellistena bifasciata
Mordellistena cervicalis
Mordellistena cf. lutea
Mordellistena ornata
Mordellistena picilabris
Mordellistena sp.
Mordellochroa scapularis
Paramordellaria triloba
Tomoxia lineela
Family
Mycetophagus pluripunctatus
Family
Carpophilus sayi
Conotelus obscurus
Glischrochilus fasciatus
Glischrochilus quadrisiginatus
Glischrochilus sanguinolentus subsp. sanguinolentus
Omosita colon
Stelidota octomaculata
Family
Pedilus lugubris
Family
Acylomus pugetanus
Olibrus semistriatus
Stilbus apicialis
Family
Psephenus herricki
Family
Ptilodactyla sp.
Family
Hadrobregmus notatus
Family
Dendroides canadensis
Schizotus cervicalis
Family
Pelecotoma flavipes
Ripiphorus fasciatus
Family
Amphimallon majale
Aphodius granarius
Ataenius strigatus
Calamosternus granarius
Onthophagus orpheus subsp. canadensis
Osmoderma scabra
Phyllophaga futilis
Phyllophaga rugosa
Popillia japonica
Rhyssemus germanus
Trox scabrosus
Family
Cyphon laevipennis
Cyphon obscurus
Cyphon pusillus
Scirtes tibialis
Family
Hylurgopinus rufipes
Scolytus mali
Xyleborus sayi
Family
Anaspis rufa
Canifa pallipes
Family
Necrophila americana
Nicrophorus orbicollis
Oiceoptoma inaequale
Family
Ahasverus advena
Silvanus bidentatus
Telephanus velox
Family
Amischa analis
Anotylus insecatus
Anotylus tetracarinatus
Atheta brunneipennis
Bisnius blandus
Carpelimus fuliginosus
Coproporus ventriculus
Lordithon appalachianus
Lordithon cinctus
Meronera venustula
Myllaena arcana
Philonthus caeruleipennis
Philonthus flavibasis
Platydracus cinnamopterus
Scaphidium quadriguttatum
Sepedophilus cinctulus
Sepedophilus testaceus
Stenichnus scutellaris
Tachinus corticinus
Tachyporus atriceps
Tachyporus chrysomelinus
Tachyporus elegans
Tachyporus nitidulus
Trichophya pilicornis
Xantholinus linearis
Family
Cephaloon lepturoides
Family
Mallodrya subaenea
Synchroa punctata
Family
Eustrophus tomentosus
Family
Aulonothroscus constrictor
Aulonothroscus distans
Aulonothroscus sp.
Trixagus carinicollis
Trixagus chevrolati
Family
Tenebroides corticalis
Order
Family
Forficula auricularia
Forficula auricularia-A
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The second of five checklists for Kingdom Animalia, this checklist contains members of Phylum Arthopoda, Class Insecta (Orders Diptera, Ephemeroptera, and Hemiptera).
Phylum
Kingdom
Class
Order
Family
Agromyza frontella
Aulagromyza luteoscutellata
Calycomyza majuscula
Cerodontha biseta
Cerodontha dorsalis
Cerodontha fasciata
Cerodontha muscina
Chromatomyia lactuca
Japanagromyza viridula
Liriomyza brassicae
Liriomyza fricki
Nemorimyza posticata
Ophiomyia labiatarum
Ophiomyia nasuta
Ophiomyia quinta
Ophiomyia similata
Phytoliriomyza dorsata
Phytoliriomyza robiniae
Phytomyza flavicornis
Phytomyza solidaginophaga
Pseudonapomyza europaea
Family
Sylvicola alternatus
Sylvicola fuscatus
Family
Anthomyia pluvialis
Delia antiqua
Delia platura
Eustalomyia festiva
Eutrichota pilimana
Hylemyza partita
Pegomya flavifrons
Family
Mumetopia occipitalis
Stiphrosoma balteatum
Family
Dioctria baumhaueri
Efferia aestuans
Efferia albibarbis
Laphria canis
Laphria canis complex
Laphria cinerea
Laphria flavicollis
Laphria janus
Laphria sicula
Laphria thoracica
Machimus sadyates
Family
Aulacigaster neoleucopeza
Family
Anthrax irroratus
Bombylius major
Hemipenthes morio
Hemipenthes sinuosa
Hemipenthes webberi
Xenox tigrinus
Family
Calliphora livida
Cynomya cadaverina
Phormia regina
Pollenia angustigena
Pollenia griseotomentosa
Pollenia labialis
Pollenia pediculata
Pollenia rudis
Family
Asteromyia carbonifera
Asteromyia laeviana
Asteromyia modesta
Asteromyia tumifica
Janetiella glechomae
Mayetiola destructor
Family
Stilobezzia antennalis
Family
Bryophaenocladius ictericus
Bryophaenocladius sp. 8ES
Camptocladius stercorarius
Chironomus acidophilus
Chironomus dilutus
Chironomus maturus
Chironomus melanescens
Chironomus ochreatus
Cladotanytarsus atridorsum
Conchapelopia telema
Corynoneura scutellata
Cricotopus annulator cmplx
Cricotopus bicinctus
Cricotopus sp. 18ES
Cricotopus sp. 19ES
Cricotopus tremulus
Cricotopus triannulatus
Cricotopus trifascia
Cricotopus vierriensis
Dicrotendipes modestus
Dicrotendipes tritomus
Gymnometriocnemus brumalis
Lauterborniella agrayloides
Limnophyes natalensis
Limnophyes sp. 14ES
Metriocnemus sp. 4ES
Micropsectra nigripila
Micropsectra subletteorum
Microtendipes pedellus
Monopelopia tenuicalcar
Nanocladius anderseni
Nilotanypus fimbriatus
Orthocladius carlatus
Orthocladius dorenus
Orthocladius oliveri
Orthocladius rivulorum
Pagastia orthogonia
Paraphaenocladius impensus
Paratanytarsus dissimilis
Paratanytarsus grimmii
Paratanytarsus laccophilus
Paratanytarsus sp. 7TE
Paratanytarsus sp. TE03
Polypedilum convictum
Psectrocladius obvius
Rheocricotopus robacki
Rheotanytarsus pellucidus
Smittia edwardsi
Smittia sp. 14ES
Smittia sp. 22ES
Smittia sp. 23ES
Smittia sp. 8ES
Stempellinella fimbriata
Tanytarsus glabrescens
Tanytarsus guerlus
Tanytarsus mendax
Tanytarsus recurvatus
Tanytarsus wirthi
Thienemanniella xena
Family
Chaetochlorops inquilinus
Elachiptera costata
Elachiptera nigriceps
Elachiptera sibirica
Eribolus longulus
Gaurax dubius
Gaurax pallidipes
Gaurax varihalteratus
Hapleginella conicola
Malloewia abdominalis
Malloewia nigripalpis
Olcella provocans
Oscinella frit
Oscinisoma alienum
Psilacrum arpidia
Rhopalopterum carbonarium
Thaumatomyia glabra
Family
Gymnochiromyia concolor
Family
Clusia czernyi
Clusia lateralis
Clusiodes johnsoni
Clusiodes melanostomus
Sobarocephala flaviseta
Sobarocephala setipes
Family
Physocephala marginata
Family
Aedes canadensis
Aedes cinereus
Aedes sp.
Aedes stimulans
Aedes vexans
Anopheles quadrimaculatus
Coquillettidia perturbans
Culex restuans
Culex territans
Mansonia perturbans
Family
Dolichopus orichalceus
Dolichopus terminalis
Gymnopternus celer
Medetera signaticornis
Neurigona disjuncta
Xanthochlorus helvinus
Family
Chymomyza amoena
Drosophila affinis
Drosophila falleni
Leucophenga varia
Scaptomyza adusta
Family
Rhamphomyia versicolor
Family
Athyroglossa dinorata
Athyroglossa granulosa
Axysta extera
Coenia curvicauda
Discocerina obscurella
Discomyza incurva
Hyadina albovenosa
Hydrellia albilabris
Hydrellia griseola
Hydrellia ischiaca
Hydrellia notata
Nostima scutellaris
Ochthera anatolikos
Parydra aquila
Philygria debilis
Philygria obtecta
Pseudohyadina longicornis
Scatella favillacea
Scatella stagnalis
Scatella tenuicosta
Scatophila despecta
Scatophila virildella
Family
Fannia armata
Family
Suillia quinquepunctata
Family
Leptopeza flavipes
Platypalpus annulatus
Platypalpus holosericus
Platypalpus melleus
Platypalpus niger
Platypalpus pulicarius
Platypalpus stabilis
Platypalpus unguiculatus
Tachydromia aemula
Family
Orfelia nemoralis
Family
Lauxania shewelli
Poecilominettia puncticeps
Family
Epiphragma fasciapenne
Erioptera caliptera
Erioptera ebenina
Helius flavipes
Ilisia venusta
Ormosia affinis
Ormosia meigenii
Pseudolimnophila inornata
Family
Lonchoptera bifurcata
Family
Compsobata univitta
Rainieria antennaepes
Family
Leptometopa latipes
Paramyia nitens
Pholeomyia indecora
Family
Coenosia tigrina
Helina depuncta
Helina evecta
Helina rufitibia
Lispe albitarsis
Lispocephala erythrocera
Macrorchis ausoba
Muscina levida
Myospila meditabunda
Family
Aglaomyia gatineau
Exechia attrita
Mycetophila caudata
Mycetophila fungorum
Mycetophila ocellus
Paratinia recurva
Symmerus lautus
Trichonta submaculata
Zygomyia zaitzevi
Family
Odinia betulae
Odinia meijerei
Family
Geomyza apicalis
Geomyza tripunctata
Family
Pedicia inconstans
Family
Conicera dauci
Megaselia arcticae
Megaselia citrinella
Megaselia fungivora
Megaselia lucifrons
Megaselia nigriceps
Megaselia rufipes
Megaselia variana
Family
Pipunculus hertzogi
Family
Loxocera cylindrica
Psila lateralis
Psila persimilis
Psila rosae
Family
Psychoda trinodulosa
Family
Bolbomyia nana
Chrysopilus thoracicus
Rhagio tringarius
Family
Boettcheria bisetosa
Boettcheria cimbicis
Sarcophaga subvicina
Senotainia trilineata
Family
Americina adusta
Megaphthalma pallida
Parallelomma vittatum
Scathophaga furcata
Family
Bradysia difformis
Bradysia fenestralis
Bradysia nitidicollis
Bradysia pallipes
Bradysia scabricornis
Bradysia vagans
Camptochaeta uniformis
Corynoptera bicuspidata
Corynoptera cuniculata
Corynoptera melanochaeta
Corynoptera saccata
Corynoptera subcavipes
Cratyna ambigua
Ctenosciara hyalipennis
Leptosciarella scutellata
Lycoriella castanescens
Lycoriella perochaeta
Lycoriella stylata
Scatopsciara atomaria
Sciara humeralis
Family
Poecilographa decorum
Pteromicra similis
Tetanocera plumosa
Trypetoptera canadensis
Family
Nemopoda nitidula
Saltella sphondylii
Sepsis punctum
Family
Prosimulium arvum
Prosimulium mixtum
Simulium vittatum
Family
Apteromyia claviventris
Coproica acutangula
Coproica ferruginata
Coproica hirtula
Copromyza equina
Gonioneura spinipennis
Ischiolepta pusilla
Leptocera caenosa
Leptocera erythrocera
Lotophila atra
Minilimosina fungicola
Minilimosina intercepta
Opalimosina mirabilis
Pullimosina heteroneura
Pullimosina longicosta
Pullimosina pullula
Rachispoda fumipennis-group
Rachispoda limosa
Rachispoda lutosa-group
Sclerocoelus sordipes
Spelobia bifrons
Spelobia clunipes
Spelobia luteilabris
Spelobia ochripes
Spelobia semioculata
Sphaerocera curvipes
Telomerina flavipes
Trachyopella nuda
Family
Actina viridis
Allognosta fuscitarsis
Allognosta obscuriventris
Caloparyphus tetraspilus
Euparyphus stigmaticalis
Microchrysa polita
Nemotelus bruesii
Nemotelus centralis
Psellidotus meganticus
Ptecticus gigliotosi
Sargus decorus
Stratiomys norma
Stratiomys obesa
Family
Allograpta micrura
Allograpta obliqua
Brachyopa sedmani
Brachypalpus oarus
Chalcosyrphus libo
Chalcosyrphus nemorum
Chrysotoxum pubescens
Dasysyrphus venustus
Epistrophe nitidicollis
Eristalis arbustorum
Eristalis dimidiata
Eristalis flavipes
Eristalis stipator
Eristalis tenax
Eristalis transversa
Eumerus sp.
Eupeodes americanus
Eupeodes volucris
Ferdinandea buccata
Helophilus fasciatus
Heringia salax
Lejota aerea
Mallota posticata
Melanotstoma mellinum
Merodon equestris
Microdon tristis
Myolepta nigra
Neoascia distincta
Ocyptamus fuscipennis
Orthonevra nitida
Paragus haemorrhous
Parhelophilus laetus
Platycheirus hyperboreus
Platycheirus obscurus
Platycheirus quadratus
Platycheirus scambus
Sericomyia chrysotoxoides
Sphaerophoria asymmetrica
Sphaerophoria bifurcata
Sphaerophoria brevipilosa
Sphaerophoria contigua
Sphaerophoria philanthus
Sphegina keeniana
Sphegina petiolata
Spilomyia longicornis
Syritta pipiens
Syrphus rectus
Syrphus ribesii
Syrphus torvus
Toxomerus geminatus
Toxomerus marginatus
Xanthogramma flavipes
Xylota quadrimaculata
Family
Chrysops aestuans
Chrysops ater
Chrysops calvus
Chrysops lateralis
Chrysops striatus
Chrysops vittatus
Hybomitra epistates
Hybomitra lasiophthalma
Tabanus lineola
Family
Actia interrupta
Blepharomyia pagana
Campylocheta teliosis
Ceracia dentata
Homalactia harringtoni
Leschenaultia exul
Lydina americana
Oswaldia minor
Phorocera obscura
Siphona hokkaidensis
Siphona intrudens
Siphona pisinnia
Family
Euaresta bella
Eurosta solidaginis
Eutreta novaeboracensis
Rhagoletis suavis
Urophora cardui
Xanthomyia platyptera
Family
Nephrotoma cornicina
Tipula (Beringotipula) coloradensis
Tipula dorsimacula
Tipula mallochi
Family
Macroceromys terminalis
Solva pallipes
Xylomyia americana
Xylomyia simillima
Xylomyia tenthredinoides
Family
Xylophagus lugens
Xylophagus reflectens
Order
Family
Baetis intercalaris
Callibaetis ferrugineus
Cloeon dipterum
Iswaeon anoka
Family
Caenis latipennis
Family
Hexagenia limbata
Family
Stenacron interpunctatum
Order
Family
Cardiastethus borealis
Orius insidiosus
Orius tristicolor
Family
Acyrthosiphon malvae
Acyrthosiphon pisum
Aphis glycines
Aphis middletonii
Aphis rubicola
Eriosoma americanum
Eucallipterus tiliae
Lipaphis pseudobrassicae
Melaphis rhois
Rhopalosiphum nymphaeae
Schizaphis scirpicola
Family
Belostoma flumineum
Family
Neoneides muticus
Family
Aphrophora alni
Aphrophora cribrata
Clastoptera obtusa
Lepyronia quadrangularis
Neophilaenus lineatus
Philaenus spumarius subsp. quadrimaculatus
Philaenus spumarius
Family
Agallia quadripunctata
Agallia sp.
Agalliopsis ancistra
Agalliopsis sp.
Anoscopus flavostriatus
Anoscopus flavostrigata
Aphrodes sp.
Arboridia sp.
Athysansus argentarius
Balclutha sp.
Ceratagallia sp.
Chlorotettix unicolor
Cicadula melanogaster
Colladonus clitellarius
Cosmotettix cf. bilineatus
Cuerna striata
Deltocephalus pulicaris
Dicraneura sp.
Dikraneura mali
Dikrella cruentata
Diplocolenus abdominalis
Doratura stylata
Draeculacephala constricta
Draeculacephala sp.
Elymana caduca
Empoasca coccinea
Empoasca decipiens
Empoasca fabae
Empoasca sp.
Erasmoneura nigra
Eratoneura certa
Eratoneura flexibilis
Errastunus ocellaris
Erythridula dunni
Erythridula scytha
Erythridula tenuispica
Erythridula wysongi
Erythroneura aza
Erythroneura bakeri
Erythroneura elegans
Erythroneura ontari
Erythroneura ontari
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The third of five checklists for Kingdom Animalia, this checklist contains members of Phylum Arthopoda, Class Insecta (Orders Hemiptera, Hymenoptera and Lepidoptera).
Class
Order
Family
Erythroneura rubrella
Erythroneura sp.
Erythroneura tricincta
Erythroneura vitifex
Erythroneura vitis
Erythroneura vulnerata
Eupteryx atropunctata
Eupteryx flavoscuta
Fieberiella florii
Forcipata acclina
Forcipata loca
Graphocephala coccinea
Graphocephala sp.
Gyponana sp.
Hymetta balteata
Jikradia olitoria
Latalus ocellaris
Macropsis basalis
Macrosteles quadrilineatus
Macrosteles sp.
Macrosteles variatus
Neokolla hieroglyphica
Oncopsis sobria
Osbornellus limosus
Penthimia americana
Scaphoideus major
Scaphoideus sp.
Scaphytopius sp.
Sorhoanus pascuellus
Typhlocyba arsinoe
Typhlocyba niobe
Typhlocyba pomaria
Zonocyba hockingensis
Family
Tibicen canicularis
Family
Clastoptera proteus
Family
Palmacorixa buenoi
Sehirus cinctus
Trichocorixa borealis
Trichocorixa sexcincta
Family
Sehirus cinctus cinctus
Family
Cymus angustatus
Family
Delphacodes sp.
Javasella pellucida
Kosswigianella lutulenta
Liburniella ornata
Megamelus lunatus
Pissonotus basalis
Family
Cedusa incisa
Family
Scolops sulcipes
Family
Metcalfa pruinosa
Family
Gelastocoris oculatus
Family
Aquarius remigis
Family
Acanalonia bivittata
Family
Kleidocerys resedae subsp. geminatus
Lygaeus kalmii
Nysius sp.
Family
Atymna helena
Campylenchia latipes
Ceresa sp.
Cyrtolobus sp.
Enchenopa binotata
Entylia carinata
Micrutalis calva
Publilia concava
Telamona decorata
Family
Adelphocoris lineolatus
Amblytylus nasutus
Capsus ater
Ceratocapsus sp.
Chlamydatus associatus
Chlamydatus sp.
Collaria meilleurii
Criocoris saliens
Deraeocoris sp.
Fulvius slateri
Halticus sp.
Heterocordylus malinus
Leptopterna dolabrata
Litomiris sp.
Lopidea sp.
Lygidea sp.
Lygocoris pabulinus
Lygus sp.
Nabicula subcoleoptrata
Nabis roseipennis
Neolygus sp.
Orthocephalus sp.
Orthops scutellatus
Pagasa fusca
Paraproba capitata
Phoenicocoris strobicola
Phytocoris sp.
Pithanus maerkelii
Plagiognathus sp.
Poecilocapsus lineatus
Prepops sp.
Slaterocoris sp.
Stenodema trispinosa
Stenodema vicinum
Stenotus binotatus
Taedia sp.
Family
Hoplistoscelis sordidus
Nabis rufusculus
Family
Notonecta undulata
Family
Crophius disconotus
Family
Phlegyas abbreviatus
Family
Acrosternum hilare
Banasa calva
Banasa dimidiata
Brochymena quadripustulata
Cosmopepla lintneriana
Euschistus tristigmus subsp. luridus
Neottiglossa undata
Parabrochymena arborea
Picromerus bidens
Podisus maculiventris
Family
Phymata americana
Piesma cinerum
Family
Pachypsylla celtidismamma
Psyllopsis fraxinicola
Family
Empicornis errabundus
Sinea diadema
Zelus luridus
Zelus tetracanthus
Family
Arhyssus sp.
Harmostes reflexulus
Stictopleurus punctiventris
Family
Ligyrocoris diffusus
Megalonotus sabulicola
Sphragisticus nebulosus
Family
Saldula confluenta
Saldula pallipes
Saldula saltatoria
Family
Corimelaena pulicaria
Galgupha atra
Family
Corythucha marmorata
Corythucha sp.
Dictyla echii
Order
Family
Andrena andrenoides
Andrena barbilabris
Andrena canadensis
Andrena carlini
Andrena erythrogaster
Andrena hirticincta
Andrena nasonii
Andrena obscuripennis
Andrena rudbeckiae
Andrena solidaginis
Andrena sp.
Andrena thaspii
Andrena wilkella
Perdita octomaculata
Pseudopanurgus nebrascensis
Family
Anthophora furcata
Ceratina calcarata
Ceratina dupla
Ceratina sp.
Melissodes desponsa
Family
Apis mellifera
Bombus bimaculatus
Bombus borealis
Bombus griseocollis
Bombus impatiens
Bombus perplexus
Bombus rufocinctus
Bombus vagans
Ceratina mikmaqi
Melissodes boltoniae
Melissodes communis
Melissodes druriella
Melissodes illata
Melissodes subillata
Melissodes trinodis
Melissodes wheeleri
Nomada articulata
Nomada bella
Nomada bethunei
Nomada cressonii
Nomada luteoloides
Nomada pygmaea
Nomada subrutila
Triepeolus helianthi
Triepeolus nigrihirtus
Triepeolus obliteratus
Triepeolus simplex
Xylocopa virginiana
Family
Parasierola sp.
Family
Aphidius ervi
Ascogaster quadridentata
Asobara rufescens
Asobara sp.
Cotesia xylina
Diaeretiella rapae
Diolcogaster facetosa
Ephedrus lacertosus
Peristenus sp.
Pholetesor ornigis
Pygostolus falcatus
Spathius elegans
Family
Conura albifrons
Family
Cleptes semiauratus
Family
Colletes eulophi
Colletes hyalinus
Colletes mandibularis
Colletes nudus
Colletes simulans
Hylaeus affinis
Hylaeus cf. affinis
Hylaeus ellipticus
Hylaeus mesillae subsp. cressoni
Hylaeus modestus
Hylaeus sp.
Family
Astata unicolor
Cerceris arelate
Cerceris atramontensis
Cerceris clypeata
Crossocerus annulipes
Crossocerus barbipes
Crossocerus elongatulus
Crossocerus tarsatus subsp. planipes
Diodontus minutus
Ectemnius cephalotes
Ectemnius continuus
Ectemnius lapidarius
Ectemnius maculosus
Ectemnius stirpicola
Gorytes atricornis
Gorytes simillimus
Hoplisoides nebulosus
Lestica confluenta
Lestica producticollis
Lyroda subita
Mimesa pauper
Oxybelus uniglumis
Passaloecus cuspidatus
Passaloecus singularis
Pemphredon inornata
Pemphredon lethifer
Philanthus bilunatus
Pison koreense
Psen monticola
Rhopalum coarctatum
Saygorytes phaleratus
Solierella peckhami
Stigmus americanus
Stigmus fraternus
Tachysphex antennatus
Tachysphex pompiliformis
Trypoxylon attenuatum
Trypoxylon carinatum
Trypoxylon clavicerum
Trypoxylon frigidum
Trypoxylon johnsoni
Trypoxylon lactitarse
Trypoxylon politum
Family
Belyta validicornis
Family
Copidosoma floridanum
Family
Eupelmus vesicularis
Family
Camponotus herculeanus
Camponotus nearcticus
Camponotus pennsylvanicus
Camponotus sp.
Crematogaster cerasi
Crematogaster sp.
Formica lasioides
Formica sp.
Lasius alienus
Lasius claviger
Lasius nearcticus
Lasius neoniger
Lasius sp.
Leptothorax ambiguus
Myrmica sp.
Ponera pennsylvanica
Prenolepis imparis
Stenamma brevicorne
Stigmatomma pallipes
Tapinoma sessile
Temnothorax ambiguus
Family
Agapostemon radiatus
Agapostemon sericeus
Agapostemon virescens
Augochlora aurata
Augochlora pura
Augochlorella aurata
Dialictus sp.
Halictus confusus
Halictus ligatus
Halictus rubicundus
Lasioglossum anomalum
Lasioglossum atwoodi
Lasioglossum birkmanni
Lasioglossum bruneri
Lasioglossum comagenense
Lasioglossum coriaceum
Lasioglossum cressonii
Lasioglossum dreisbachi
Lasioglossum ephialtum
Lasioglossum fattigi
Lasioglossum foxii
Lasioglossum heterognathum
Lasioglossum hitchensi
Lasioglossum imitatum
Lasioglossum leucozonium
Lasioglossum lineatulum
Lasioglossum macoupinense
Lasioglossum michiganense
Lasioglossum nigroviride
Lasioglossum paradmirandum
Lasioglossum pectinatum
Lasioglossum pectorale
Lasioglossum perpunctatum
Lasioglossum pilosum
Lasioglossum platyparium
Lasioglossum pruinosum
Lasioglossum sp. 1
Lasioglossum sp. 2
Lasioglossum subversans
Lasioglossum tegulare
Lasioglossum tenax
Lasioglossum timothyi
Lasioglossum versans
Lasioglossum versatum
Lasioglossum vierecki
Lasioglossum weemsi
Lasioglossum zonulum
Lasioglossum zophops
Sphecodes atlantis
Sphecodes clematidis
Sphecodes confertus
Sphecodes cressonii
Sphecodes davisii
Sphecodes dichrous
Sphecodes heraclei
Sphecodes levis
Sphecodes minor
Sphecodes persimilis
Sphecodes pycnanthemi
Sphecodes ranunculi
Sphecodes sp.
Sphecodes stygius
Sphecodes wheeleri
Family
Agrothereutes abbreviatus
Agrypon flexorium
Aritranis director
Bathyplectes anurus
Bathythrix decipiens
Campoletis flavicincta
Cryptus albitarsis
Cylloceria melancholica
Cymodusa distincta
Diadegma pendulum
Dialipsis dissimilis
Diplazon laetatorius
Dolichomitus irritator
Dusona minor
Enytus apostata
Exochus nigripalpis
Hyposoter inquinatus
Ichneumon discoensis
Ischnus inquisitorius
Iseropus stercorator
Lissonota coracina
Megacara hortulana
Mesochorus suomiensis
Ophion bilineatus
Ophion clave
Ophion idoneus
Ophion sp. 5 MDS2014
Oresbius taeniatus
Orthocentrus fulvipes
Phobocampe bicingulata
Pimpla aequalis
Pleolophus basizonus
Podoschistus vittifrons
Stenomacrus nemoralis
Tranosema rostrale
Zaglyptus varipes
Family
Leucospis affinis
Family
Anthidiellum notatum
Anthidium manicatum
Coelioxys rufitarsus
Heriades variolosa subsp. variolosa
Hoplitis pilosifrons
Hoplitis producta
Hoplitis truncata
Megachile brevis
Megachile campanulae
Megachile frigide
Megachile gemula
Megachile inermis
Megachile latimanus
Megachile lippiae
Megachile mendica
Megachile montivaga
Megachile perihirta
Megachile pugnata
Megachile relativa
Megachile rotundata
Megachile texana
Osmia coerulescens
Osmia conjuncta
Osmia lignaria
Osmia proxima
Osmia pumila
Stelis lateralis
Family
Myrmosa unicolor
Pseudomethoca frigida
Family
Anaphes listronoti
Gonatocerus morrilli
Ooctonus silvensis
Family
Platygaster variabilis
Synopeas pennsylvanicum
Telenomus podisi
Family
Agenioideus cinctellus
Agenioideus humilis
Anoplius imbellis
Anoplius nigerrimus
Anoplius virginiensis
Aporinellus wheeleri
Arachnospila michiganensis subsp. michiganensis
Auplopus carbonarius
Auplopus mellipes subsp. variitarsatus
Auplopus nigrellus
Caliadurgus fasciatellus subsp. alienatus
Dipogon sayi subsp. sayi
Episyron biguttatus subsp. biguttatus
Priocnemis cornica
Priocnemis germana
Priocnemis minorata
Priocnemis notha subsp. notha
Priocnemis scitula subsp. relicta
Family
Mesopolobus bruchophagi
Family
Sapyga centrata
Family
Sierolomorpha canadensis
Family
Tremex columba
Family
Chalybion californicum
Isodontia auripes
Isodontia mexicana
Sceliphron caementarium
Sphex ichneumoneus
Family
Ametastegia aperta
Ametastegia pallipes
Caulocampus acericaulis
Dolerus asper
Dolerus nitens
Empria maculata
Empria nordica
Fenusa ulmi
Halidamia affinis
Macrophya flavolineata
Metallus lanceolatus
Monophadnus pallescens
Pachynematus extensicornis
Periclista sp. tM8
Priophorus compressicornis
Pristiphora chlorea
Taxonus epicera
Taxonus pallicoxus
Taxonus terminalis
Tomostethus multicinctus
Family
Trichogramma platneri
Family
Ancistrocerus adiabatus subsp. adiabatus
Ancistrocerus albophaleratus
Ancistrocerus antilope subsp. antilope
Ancistrocerus campestris
Ancistrocerus catskill
Ancistrocerus unifasciatus subsp. unifasciatus
Dolichovespula arenaria
Eumenes crucifer
Eumenes fraternus
Euodynerus foraminatus subsp. foraminatus
Euodynerus leucomelas subsp. leucomelas
Monobia quadridens
Parancistrocerus leionotus
Parancistrocerus pedestris
Parancistrocerus pensylvanicus
Parancistrocerus pensylvanicus subsp. pensylvanicus
Polistes dominula
Polistes fuscatus
Symmorphus canadensis
Symmorphus cristatus
Vespula flavopilosa
Vespula germanica
Vespula maculifrons
Vespula vidua
Vespula vulgaris
Order
Family
Asaphocrita busckiella
Blastobasis glandulella
Family
Bucculatrix ainsliella
Bucculatrix pomifoliella
Family
Cosmopterix montisella
Teladoma helianthi
Family
Acentria ephemerella
Agriphila vulgivagella
Anania funebris
Desmia maculalis
Elophila gyralis
Elophila icciusalis
Elophila tinealis
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The fourth of five checklists for Kingdom Animalia, this checklist contains members of Phylum Arthopoda, Class Insecta (Orders Lepidoptera to Trichoptera), and Class Malacostraca. From Phylum Chordata, it contains Classes Actinopterygii, Amphibia, and Aves (Orders Anseriformes to Passeriformes).
Kingdom
Phylum
Class
Order
Family
Loxostege sticticalis
Microcrambus elegans
Nomophila nearctica
Perispasta caeculalis
Petrophila bifascialis
Sitochroa palealis
Udea rubigalis
Urola nivalis
Family
Agonopterix arenella
Agonopterix pulvipennella
Depressaria depressana
Machimia tentoriferella
Family
Drepana arcuata
Family
Perittia herrichiella
Family
Epermenia albapunctella
Family
Apantesis phalerata
Cisseps fulvicollis
Ctenucha virginica
Hypena madefactalis
Hypena scabra
Hyphantria cunea
Lymantria dispar subsp. dispar
Rivula propinqualis
Family
Bryotropha hodgesi
Chionodes fondella
Chrysoesthia sexguttella
Dichomeris furia
Dichomeris inserrata
Dichomeris leuconotella
Dichomeris ligulella
Dichomeris mercatrix
Helcystogramma hystricella
Metzneria lappella
Monochroa fragariae
Scrobipalpa acuminatella
Scrobipalpula physaliella
Scrobipalpula sacculicola
Sinoe chambersi
Xenolechia ontariensis
Family
Alsophila pometaria
Besma quercivoraria
Biston betularia
Campaea perlata
Coryphista meadii
Costaconvexa centrostrigaria
Ecliptopera silaceata
Ennomos magnaria
Epirrhoe alternata
Euchlaena serrata
Idaea dimidiata
Operophtera bruceata
Orthonama obstipata
Phigalia titea
Plagodis phlogosaria
Pleuroprucha insulsaria
Scopula inductata
Speranza pustularia
Synchlora frondaria
Trichodezia albovittata
Xanthorhoe ferrugata
Xanthorhoe lacustrata
Family
Acrocercops astericola
Caloptilia packardella
Cameraria saccharella
Cremastobombycia solidaginis
Parornix betulae
Parornix crataegifoliella
Phyllocnistis ampelopsiella
Phyllocnistis vitegenella
Phyllonorycter clemensella
Phyllonorycter maestingella
Phyllonorycter ostryaefoliella
Phyllonorycter propinquinella
Phyllonorycter salicifoliella
Phyllonorycter trinotella
Phyllonorycter tritaenianella
Family
Anatrytone logan
Ancyloxypha numitor
Atalopedes campestris
Carterocephalus palaemon
Epargyreus clarus
Erynnis baptisiae
Erynnis juvenalis
Erynnis lucilius
Euphyes conspicua
Euphyes dion
Euphyes vestris
Pholisora catullus
Poanes hobomok
Poanes massasoit
Poanes viator
Polites mystic
Polites origenes
Polites peckius
Polites themistocles
Pompeius verna
Thymelicus lineola
Wallengrenia egeremet
Family
Malacosoma disstria
Tolype velleda
Family
Celastrina ladon
Celastrina neglecta
Cupido comyntas
Feniseca tarquinius
Lycaena hyllus
Satyirum titus
Satyrium acadicum
Satyrium calanus
Satyrium caryaevorus
Satyrium liparops
Family
Dasychira basiflava
Orgyia definita
Orgyia leucostigma
Family
Mompha terminella
Family
Ectoedemia argyropeza
Stigmella microtheriella
Stigmella rhamnicola
Family
Achatia distincta
Agrotis ipsilon
Agrotis venerabilis
Allagrapha aerea
Amphipoea americana
Amphipoea interoceanica
Amphipyra pyramidoides
Anathix ralla
Apamea devastator
Autographa precationis
Caenurgina crassiuscula
Catocala cerogama
Catocala grynea
Cerastis tenebrifera
Chrysodeixis includens
Crocigrapha normani
Cucullia asteroides
Cucullia convexipennis
Euplexia benesimilis
Eupsilia devia
Feltia jaculifera
Hyppa xylinoides
Idia aemula
Lacinipolia meditata
Lacinipolia renigera
Leucania commoides
Leucania multilinea
Leucania phragmitidicola
Leucania pseudargyria
Loscopia velata
Macrochilo absorptalis
Melanchra adjuncta
Meropleon diversicolor
Morrisonia confusa
Mythimna unipuncta
Noctua pronuba
Ochropleura implecta
Oligia modica
Orthosia hibisci
Orthosia rubescens
Palthis angulalis
Peridroma saucia
Phalaenostola metonalis
Protodeltote albidula
Pseudohermonassa bicarnea
Renia adspergillus
Striacosta albicosta
Sunira bicolorago
Xestia smithii
Family
Nola ovilla
Family
Schizura unicornis
Symmerista leucitys
Family
Aglais milberti
Asterocampa clyton
Boloria Bellona
Boloria selene
Cercyonis pegala
Chlosyne nycteis
Coenonympha tullia
Danaus plexippus
Euphydryas phaeton
Euptoieta claudia
Junonia coenia
Lethe anthedon
Lethe appalachia
Lethe eurydice
Libytheana carinenta
Limenitis archippus
Limenitis arthemis
Megisto cymela
Nymphalis antiopa
Nymphalis l-album
Phyciodes cocyta
Phyciodes tharos
Polygonia comma
Polygonia interrogationis
Polygonia progne
Speyeria Cybele
Vanessa atalanta
Vanessa cardui
Vanessa virginiensis
Family
Papilio canadensis
Papilio cresphontes
Papilio glaucus
Papilio polyxenes
Family
Colias eurytheme
Colias philodice
Pieris oleracea
Pieris rapae
Pyrisitia lisa
Family
Plutella porrectella
Plutella xylostella
Family
Psyche casta
Family
Gillmeria pallidactyla
Hellinsia homodactylus
Hellinsia pectodactylus
Family
Deidamia inscriptum
Family
Coptotriche badiiella
Family
Acleris chalybeana
Acleris cornana
Ancylis muricana
Argyrotaenia mariana
Choristoneura rosaceana
Cochylis hoffmanana
Cochylis temerana
Endothenia hebesana
Epinotia medioviridana
Eucosma similana
Grapholita prunivora
Olethreutes atrodentana
Olethreutes fasciatana
Olethreutes permundana
Pandemis lamprosana
Phaneta ochrocephala
Phaneta parmatana
Phaneta tomonana
Platynota idaeusalis
Pristerognatha fuligana
Proteoteras aesculana
Order
Family
Mantis religiosa subsp. religiosa
Order
Family
Bittacus strigosus
Family
Panorpa galerita
Panorpa latipennis
Panorpa subfurcata
Order
Family
Chrysopa oculata
Family
Hemerobius humulinus
Hemerobius stigma
Micromus posticus
Order
Family
Aeshna umbrosa
Anax junius
Family
Hetaerina americana
Family
Argia moesta
Enallagma antennatum
Enallagma civile
Enallagma ebrium
Enallagma exsulans
Enallagma geminatum
Enallagma signatum
Ischnura kellicotti
Ischnura posita
Ishnura verticalis
Nehalennia irene
Family
Lestes disjunctus
Lestes rectangularis
Family
Erythemis simplicicollis
Libellula luctuosa
Libellula pulchella
Libellula quadrimaculata
Pachydiplax longipennis
Plathemis lydia
Sympetrum internum
Sympetrum obtrusum
Sympetrum semicinctum
Tramea lacerat
Order
Family
Cholealtis conspera
Chorthippus curtipennis
Chortophaga viridifasciata
Dissosteira carolina
Dissosteria carolina
Melanoplus bivatattus
Melanoplus sanguinipes subsp. sanguinipes
Melanoplus sp.
Family
Allonemobius fasciatus
Gryllus veletis
Oecanthus nigricornis
Oecanthus quadripunctatus
Family
Tetrix arenonsum subsp. angusta
Tetrix subulata
Family
Conocephalus brevipennis
Scudderia sp.
Order
Family
Diapheromera femorata
Order
Family
Sweltsa onkos
Order
Family
Polypsocus corruptus
Family
Caecillius sp.
Family
Valenzuela flavidus
Family
Ectopsocus meridionalis
Family
Mesopsocus unipunctatus
Family
Peripsocus subfasciatus
Family
Blaste opposita
Metylophorus novaescotiae
Family
Graphopsocus cruciatus
Order
Family
Aeolothrips ericae
Family
Haplothrips verbasci
Family
Chirothrips manicatus
Odontothrips biuncus
Taeniothrips inconsequens
Order
Family
Helicopsyche borealis
Family
Ceratopsyche morosa
Cheumatopsyche campyla
Hydropsyche phalerata
Family
Agraylea multipunctata
Hydroptila armata
Hydroptila perdita
Hydroptila spatulata
Orthotrichia cristata
Family
Oecetis avara
Oecetis cinerascens
Oecetis inconspicua
Oecetis nocturna
Family
Ironoquia punctatissima
Pycnopsyche antica
Family
Banksiola crotchi
Family
Plectrocnemia cinerea
Class
Order
Family
Hyalella azteca
Order
Family
Orconectes propinquus
Order
Family
Trachelipus rathkii
Family
Hyloniscus riparius
Trichoniscus pusillus
Phylum
Class
Order
Family
Catostomus catostomus
Catostomus commersonii
Hypentelium nigricans
Moxostoma erythrurum
Moxostoma valenciennesi
Family
Cyprinella spiloptera
Cyprinus Carpio
Luxilus chrysocephalus
Luxilus cornutus
Nocomis biguttatus
Nocomis micropogon
Notropis heterodon
Notropis photogenis
Pimephales notatus
Rhinichthys atratulus
Semotilus atromaculatus
Order
Family
Esox lucius
Order
Family
Culaea inconstans
Order
Family
Ambloplites rupestris
Lepomis gibbosus
Micropterus dolomieu
Micropterus salmoides
Pomoxis nigromaculatus
Family
Etheostoma blennioides
Etheostoma exile
Etheostoma nigrum
Perca flavescens
Percina maculata
Order
Family
Salvelinus fontinalis
Order
Family
Ameiurus nebulosus
Noturus flavus
Class
Order
Family
Anaxyrus americanus
Family
Hyla versicolor
Pseudacris crucifer
Pseudacris triseriata
Family
Lithobates catesbeiana
Lithobates clamitans
Lithobates pipiens
Lithobates sylvatica
Order
Family
Ambystoma jeffersonianum x laterale var. hybrid species: jefferson and blue-spotted
Ambystoma laterale
Ambystoma maculatum
Family
Hemidactylium scutatum
Plethodon cinereus
Class
Order
Family
Aix sponsa
Anas acuta
Anas americana
Anas clypeata
Anas crecca
Anas discors
Anas platyrhynchos
Anas rubripes
Anas strepera
Aythya affinis
Aythya americana
Aythya collaris
Aythya marila
Aythya valisineria
Branta bernicla
Branta canadensis
Branta hutchinsii
Bucephala albeola
Bucephala clangula
Chen caerulescens
Clangula hyemalis
Cygnus buccinator
Cygnus columbianus
Cygnus olor
Lophodytes cucullatus
Melanerpes erythrocephalus
Mergus merganser
Mergus serrator
Oxyura jamaicensis
Order
Family
Chaetura pelagica
Family
Archilochus colubris
Order
Family
Caprimulgus vociferus
Chordeiles minor
Order
Family
Charadrius semipalmatus
Charadrius vociferus
Family
Chroicocephalus philadelphia
Larus delawarensis
Larus fuscus
Larus glaucoides
Larus hyperboreus
Larus marinus
Larus smithsonianus
Larus thayeri
Mniotilta varia
Sterna hirundo
Family
Actitis macularius
Arenaria interpres
Bartramia longicauda
Calidris alpina
Calidris bairdii
Calidris melanotos
Calidris minutilla
Calidris pusilla
Gallinago delicata
Scolopax minor
Tringa flavipes
Tringa melanoleuca
Tringa solitaria
Family
Hydroprogne caspia
Order
Family
Ardea alba
Ardea herodias
Botaurus lentiginosus
Butorides virescens
Chlidonias niger
Egretta caerulea
Order
Family
Columba livia
Zenaida macroura
Order
Family
Megaceryle alcyon
Order
Family
Coccyzus americanus
Coccyzus erythropthalmus
Order
Family
Accipiter cooperii
Accipiter gentilis
Accipiter striatus
Agelaius phoeniceus
Aquila chrysaetos
Buteo jamaicensis
Buteo lagopus
Buteo lineatus
Buteo platypterus
Circus cyaneus
Haliaeetus leucocephalus
Family
Cathartes aura
Family
Falco columbarius
Falco peregrinus
Falco sparverius
Family
Pandion haliaetus
Order
Family
Colinus virginianus
Family
Bonasa umbellus
Meleagris gallopavo
Phasianus colchicus
Order
Family
Gavia immer
Order
Family
Grus canadensis
Family
Fulica americana
Porzana carolina
Rallus limicola
Order
Family
Eremophilia alpestris
Family
Bombycilla cedrorum
Family
Calcarius lapponicus
Family
Cardinalis cardinalis
Passerina cyanea
Pheucticus ludovicianus
Plectrophenax nivalis
Family
Certhia americana
Family
Corvus brachyrhynchos
Corvus corax
Cyanocitta cristata
Family
Ammodramus leconteii
Ammodramus nelsoni
Ammodramus savannarum
Junco hyemalis
Melospiza georgiana
Melospiza lincolnii
Melospiza melodia
Passerculus sandwichensis
Passerella iliaca
Pipilo erythrophthalmus
Pipilo maculatus
Pooecetes gramineus
Spizella arborea
Spizella pallida
Spizella passerina
Spizella pusilla
Zonotrichia albicollis
Zonotrichia leucophrys
Family
Carduelis flammea
Carduelis pinus
Carduelis tristis
Carpodacus mexicanus
Carpodacus purpureus
Coccothraustes vespertinus
Loxia curvirostra
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The fifth of five checklists for Kingdom Animalia, this checklist contains records from Phylum Chordata, Class Aves (Orders Passeriformes to Strigiformes), Class Mammalia, and Class Reptilia, as well as Phylum Gastropoda.
Phylum
Class
Order
Family
Loxia leucoptera
Family
Hirundo rustica
Petrochelidon pyrrhonota
Riparia riparia
Stelgidopteryx serripennis
Tachycineta bicolor
Family
Dolichonyx oryzivorus
Euphagus carolinus
Euphagus cyanocephalus
Icterus galbula
Icterus spurius
Molothrus ater
Quiscalus quiscula
Sturnella magna
Xanthocephalus xanthocephalus
Family
Lanius excubitor
Family
Dumetella carolinensis
Mimus polyglottos
Toxostoma rufum
Family
Anthus rubescens
Family
Poecile atricapillus
Family
Cardellina canadensis
Cardellina pusilla
Geothlypis philadelphia
Geothlypis trichas
Leiothlypis celata
Leiothlypis peregrina
Leiothlypis ruficapilla
Oporornis agilis
Setophaga americana
Setophaga caerulescens
Setophaga castanea
Setophaga citrina
Setophaga coronata
Setophaga discolor
Setophaga fusca
Setophaga magnolia
Setophaga palmarum
Setophaga pensylvanica
Setophaga petechia
Setophaga pinus
Setophaga ruticilla
Setophaga striata
Setophaga tigrina
Setophaga virens
Vermivora chrysoptera
Vermivora cyanoptera
Family
Passer domesticus
Family
Nycticorax nycticorax
Parkesia noveboracensis
Picoides arcticus
Seiurus aurocapilla
Family
Regulus calendula
Regulus satrapa
Family
Sitta canadensis
Sitta carolinensis
Family
Sturnus vulgaris
Family
Polioptila caerulea
Family
Piranga olivacea
Family
Cistothorus palustris
Thryothorus ludovicianus
Troglodytes aedon
Troglodytes hiemalis
Family
Catharus fuscescens
Catharus guttatus
Catharus minimus
Catharus ustulatus
Hylocichla mustelina
Sialia sialis
Turdus migratorius
Family
Contopus cooperi
Contopus virens
Empidonax alnorum
Empidonax flaviventris
Empidonax Minimus
Empidonax traillii
Myiarchus crinitus
Sayornis phoebe
Tyrannus tyrannus
Family
Vireo flavifrons
Vireo gilvus
Vireo griseus
Vireo olivaceus
Vireo philadelphicus
Vireo solitarius
Order
Family
Phalacrocorax auritus
Order
Family
Colaptes auratus
Dryocopus Pileatus
Melanerpes carolinus
Picoides pubescens
Picoides villosus
Sphyrapicus varius
Order
Family
Podiceps auritus
Podiceps grisegena
Podilymbus podiceps
Order
Family
Aegolius acadicus
Asio flammeus
Bubo virginianus
Megascops asio
Strix varia
Class
Order
Family
Odocoileus virginianus
Order
Family
Canis latrans
Vulpes vulpes
Family
Lynx rufus
Family
Mephitis mephitis
Family
Lontra canadensis
Mustela erminea
Neovison vison
Family
Procyon lotor
Order
Family
Eptesicus fuscus
Lasionycteris noctivagans
Lasiurus borealis
Lasiurus cinereus
Myotis lucifugus
Myotis septentrionalis
Order
Family
Didelphis virginiana
Order
Family
Lepus americanus
Lepus europaeus
Sylvilagus floridanus
Order
Family
Castor canadensis
Family
Microtus pennsylvanicus
Myodes gapperi
Ondatra zibethicus
Peromyscus leucopus
Peromyscus maniculatus
Family
Zapus hudsonius
Family
Erethizon dorsatus
Family
Mus musculus
Rattus norvegicus
Family
Glaucomys sabrinus
Marmota monax
Sciurus carolinensis
Tamias striatus
Tamiasciurus hudsonicus
Order
Family
Blarina brevicauda
Sorex cinereus
Sorex fumeus
Family
Condylura cristata
Class
Order
Family
Lampropeltis triangulum subsp. triangulum
Nerodia sipedon
Opheodrys vernalis
Regina septemvittata
Storeria dekayi
Storeria occipitomaculata
Thamnophis sauritus
Thamnophis sirtalis subsp. sirtalis
Order
Family
Chelydra serpentina
Family
Chrysemys picta
Phylum
Class
Family
Anguispira alternata
Family
Hawaiia minuscula
Family
Novisuccinea ovalis
Family
Vertigo bollesiana
Order
Family
Gyraulus circumstriatus
Order
Family
Physa gyrina
Order
Family
Paravitrea multidentata
Order
Family
Deroceras reticulatum
Family
Arion fuscus
Arion subfuscus
Family
Oxychilus allarius
Zonitoides arboreus
Family
Cepaea nemoralis
Cochlicopa lubrica
Family
Helicodiscus parallelus
Family
Trochulus hispidus
Family
Succinea putris
Family
Vitrina angelicae
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. This checklist contains members of Kingdom Amoebozoa, Phylum Mycetozoa.
Class
Order
Family
Lycogala epidendrum
Order
Family
Tubifera ferruginosa
Order
Family
Stemonitis axifera
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. This checklist contains records from Kingdom Fungi, including Phyla Ascomycota, Basidiomycota, and Zygomycota.
Phylum
Dictyocatenulata alba
Phylum
Class
Order
Family
Arthonia caesia
Arthonia caudata
Arthonia radiata
Class
Order
Family
Apiosporina morbosa
Order
Family
Ischnoderma resinosum
Piptoporus betulinus
Postia caesia
Postia fragilis
Family
Ganoderma applanatum
Ganoderma lucidum
Family
Gloeoporus dichrous
Grifola frondosa
Family
Bjerkandera adusta
Climacodon septentrionale
Phlebia radiata
Porotheleum fimbriatum
Family
Steccherinum ochraceum
Family
Cerrena unicolor
Favolus alveolaris
Laetiporus sulphureus
Lenzites betulina
Polyporus badius
Polyporus brumalis
Trametes elegans
Trichaptum abietinum
Tyromyces chioneus
Family
Irpex lacteus
Class
Order
Family
Mycocalicium subtile
Phaeocalicium curtisii
Order
Family
Verrucaria calkinsiana
Class
Order
Family
Trapelia involuta
Trapelia placodioides
Order
Family
Candelaria concolor
Candelariella aurella
Candelariella efflorescens
Order
Family
Acarospora glaucocarpa
Family
Cladonia ochrochlora
Family
Lecanora allophana
Lecanora allophana f. sorediata
Lecanora pulicaris
Lecanora sambuci
Lecanora symmicta
Lecanora thysanophora
Lecidella carpathica
Family
Flavoparmelia caperata
Flavopunctelia flaventior
Melanelixia subaurifera
Parmelia squarrosa
Parmelia sulcata
Punctelia rudecta
Punctelia subrudecta
Xanthoparmelia cumberlandia
Family
Peltigera evansiana
Peltigera rufescens
Family
Bilimbia sabuletorum
Family
Protoblastenia rupestris
Psora decipiens
Family
Bacidia schweinitzii
Lecania croatica
Lecania naegelii
Family
Lepraria incana
Lepraria lobificans
Order
Family
Julella fallaciosa
Order
Family
Coenogonium pineti
Family
Graphis scripta
Family
Phlyctis spp.
Family
Conotrema urceolatum
Order
Family
Buellia punctata
Buellia stillingiana
Hyperphyscia adglutinata
Phaeophyscia adiastola
Phaeophyscia orbicularis
Phaeophyscia pusilloides
Phaeophyscia rubropulchra
Physcia adscendens
Physcia aipolia
Physcia millegrana
Physcia stellaris
Physciella chloantha
Physconia detersa
Family
Caloplaca flavovirescens
Caloplaca pyracea
Xanthomendoza fallax
Xanthomendoza ulophyllodes
Xanthoria parietina
Class
Order
Family
Bulgaria inquinans
Family
Ascocoryne cylichnium
Bisporella citrina
Chlorociboria aeruginascens
Family
Chlorencoelia versiformis
Class
Order
Family
Peziza repanda
Family
Aleuria aurantia
Jafnea semitosta
Scutellinia scutellata
Scutellinia setosa
Tarzetta cupularis
Class
Order
Family
Ovicuculispora parmeliae
Family
Hypomyces lactifluorum
Order
Family
Daldinia concentrica
Hypoxylon fragiforme
Ustulina deusta
Xylaria longipes
Xylaria polymorpha
Phylum
Class
Order
Family
Agaricus arvensis
Agaricus placomyces
Calvatia gigantea
Coprinus comatus
Lepiota clypeolaria
Lepiota cristata
Leucoagaricus naucinus
Lycoperdon perlatum
Parasola plicatilis
Family
Amanita bisporigera
Family
Conocybe rugosa
Panaeolus foenisecii
Family
Gymnopilus spectabilis
Phaeomarasmius proximans
Family
Clitopilus prunulus
Clitopilus scyphoides
Entoloma abortivum
Entoloma spp.
Entoloma strictus
Leptonia incana
Family
Arrhenia epichysium
Hygrocybe coccinea
Hygrocybe flavescens
Hygrocybe miniata
Hygrocybe pratensis
Hygrocybe punicea
Hygrocybe virginea
Hygrophorus eburneus
Family
Galerina autumnalis
Hebeloma crustuliniforme
Family
Crepidotus applanatus
Crepidotus mollis
Inocybe rimosa
Inocybe spp.
Crepidotus crocophyllus
Family
Lycoperdon pyriforme
Family
Lyophyllum decastes
Family
Baeospora myosura
Clitocybula oculus
Marasmius rotula
Micromphale foetidum
Pleurocybella porrigens
Rhodocollybia butryacea
Gymnopus acervatus
Xerula furfuracea
Family
Mycena galericulata
Mycena haematopus
Mycena inclinata
Mycena leaiana
Mycena olida
Mycena osmundicola
Mycena spp.
Panellus stipticus
Xeromphalina kauffmanii
Family
Armillaria gallica
Armillaria mellea complex
Armillaria ostoyae
Flammulina velutipes
Hymenopellis limonispora
Xerula megalospora
Family
Pleurotus ostreatus
Family
Pluteus atricapillus
Pluteus atromarginatus
Pluteus cervinus
Pluteus hongoi
Pluteus longistriatus
Pluteus umbrosus
Family
Coprinellus micaceus
Coprinopsis atramentaria
Coprinus disseminatus
Psathyrella candolleana
Family
Schizophyllum commune
Family
Agrocybe praecox
Gymnopilus penetrans
Gymnopolis sapineus
Hypholoma capnoides
Hypholoma fasciculare
Hypholoma sublateritium
Pholiota aurivella
Pholiota squarrosa
Pholiota squarrosoides
Family
Clitocybe ectypoides
Clitocybe gibba
Collybia cookei
Hygrophorus langei
Hypsizygus tessulatus
Lepista irina
Lepista nuda
Omphalina spp.
Phyllotopsis nidulans
Rhodotus palmatus
Tricholoma aurantium
Tricholoma myomyces
Tricholoma terreum
Tricholoma virgatum
Xeromphalina cauticinalis
Panellus serotinus
Family
Multiclavula mucida
Order
Family
Boletinellus merulioides
Leccinum scabrum
Family
Scleroderma areolatum
Scleroderma citrinum
Scleroderma michiganense
Family
Suillus americanus
Suillus brevipes
Suillus granulatus
Suillus luteus
Order
Family
Craterellus fallax
Order
Family
Phlebia tremellosa
Order
Family
Geastrum fimbriatum
Geastrum quadrifidum
Order
Family
Ramaria spp.
Ramaria stricta var. concolor
Order
Family
Phellinus igniarius
Order
Family
Phallus duplicatus
Phallus impudicus
Phallus ravenelii
Order
Family
Loweomyces fractipes
Family
Daedaleopsis confragosa
Fomes fomentarius
Polyporus radicatus
Polyporus squamosus
Polyporus varius
Trametes gibbosa
Trametes pubescens
Trametes versicolor
Trichaptum biforme
Truncospora ohiensis
Order
Family
Hericium americanum
Hericium coralloides
Hericium erinaceus
Family
Lactarius torminosus
Russula rosacea
Russula spp.
Family
Stereum ostrea
Stereum rugosum
Stereum striatum
Family
Lentinellus ursinus
Order
Family
Boletopsis subsquamosa
Class
Order
Family
Inocybe geophylla var. geophylla
Class
Order
Family
Calocera cornea
Class
Order
Family
Exidia alba
Exidia glandulosa
Family
Tremella foliacea
Tremella reticulate
Class
Order
Family
Ustilago maydis
Phylum
Class
Order
Family
Mortierella hyalina
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The first of two checklists for Kingdom Plantae, this checklist contains records from Phyla Bryophyta, Equisetophyta, Lycopodiophyta and Magnoliophyta (Class Liliopsida and Magnoliopsida).
Kingdom
Phylum
Class
Order
Family
Bryum caespiticium
Bryum argenteum
Family
Mnium marginatum
Family
Plagiomnium cuspidatum
Order
Family
Dicranum flagellare
Dicranum montanum
Family
Ceratodon purpureus
Ditrichum spp.
Family
Fissidens adianthoides
Fissidens taxifolius
Fissidens spp.
Family
Leucobryum glaucum
Order
Family
Encalypta procera
Order
Family
Schistidium apocarpa
Schistidium rivulare
Order
Family
Hedwigia ciliata
Order
Family
Amblystegium serpens
Amblystegium varium
Campylium chrysophyllum
Campylium hispidulum
Campylium spp.
Hygroamblystegium fluviatile
Hygroamblystegium tenax
Leptodictyum riparium
Drepanocladus aduncus
Family
Anomodon attenuatus
Anomodon rostratus
Family
Brachythecium spp.
Cirriphyllum piliferum
Eurhynchium hians
Eurhynchium pulchellum
Family
Climacium americanum
Climacium dendroides
Family
Entodon seductrix
Family
Fontinalis hypnoides
Family
Rhytidiadelphus triquetrus
Family
Callicladium haldanianum
Homomallium adnatum
Hypnum curvifolium
Hypnum imponens
Hypnum lindbergii
Hypnum spp.
Platydictya subtilis
Taxiphyllum deplanatum
Family
Leskea polycarpa
Leskeella nervosa
Family
Isopterygiopsis muelleriana
Family
Thuidium recognitum
Thuidium delicatulum
Order
Family
Orthotrichum anomalum
Order
Family
Barbula convoluta
Desmatodon obtusifolius
Hymenostylium recurvirostre
Tortella tortuosa
Barbula unguiculata
Tortula ruralis
Weissia spp.
Order
Family
Leptobryum pyriforme
Class
Order
Family
Atrichum angustatum
Atrichum undulatum
Polytrichum spp.
Class
Order
Family
Sphagnum spp.
Class
Order
Family
Tetraphis pellucida
Phylum
Class
Order
Family
Equisetum arvense
Equisetum hyemale subsp. affine
Equisetum palustre
Equisetum pratense
Equisetum scirpoides
Equisetum sylvaticum
Equisetum variegatum subsp. variegatum
Phylum
Class
Family
Family
Huperzia lucidula
Order
Family
Lycopodiella inundata
Lycopodium annotinum
Lycopodium obscurum
Lycopodium ssp.
Order
Order
Family
Selaginella eclipes
Phylum
Class
Order
Family
Sagittaria latifolia
Sagittaria rigida
Family
Arisaema triphyllum
Symplocarpus foetidus
Family
Butomus umbellatus
Family
Potamogeton crispus
Stuckenia pectinata
Family
Triantha glutinosa
Order
Family
Calla palustris
Lemna minor
Wolffia borealis
Wolffia columbiana
Order
Family
Allium tricoccum
Family
Asparagus officinalis
Convallaria majalis
Maianthemum canadense
Maianthemum racemosum subsp. racemosum
Maianthemum stellatum
Family
Iris pseudacorus
Family
Cypripedium acaule
Cypripedium arietinum
Cypripedium parviflorum var. pubescens
Cypripedium reginae
Order
Family
Carex albursina
Carex aquatilis
Carex arctata
Carex atherodes
Carex backii
Carex bebbii
Carex blanda
Carex bromoides
Carex chordorrhiza
Carex comosa
Carex crinita
Carex cristatella
Carex deweyana
Carex diandra
Carex disperma
Carex eburnea
Carex foenea
Carex formosa
Carex glaucodea
Carex gracillima
Carex granularis
Carex grisea
Carex hirtifolia
Carex hitchcockiana
Carex hystericina
Carex intumescens
Carex jamesii
Carex lacustris
Carex laevivaginata
Carex lasiocarpa
Carex laxiculmis
Carex laxiflora
Carex leptalea
Carex livida
Carex lupuliformis
Carex lupulina
Carex normalis
Carex peckii
Carex pedunculata
Carex pensylvanica
Carex plantaginea
Carex platyphylla
Carex projecta
Carex pseudocyperus
Carex retrorsa
Carex rosea
Carex scabrata
Carex schweinitzii
Carex scoparia
Carex sparganioides
Carex spp.
Carex stipata
Carex stricta
Carex tetanica
Carex tonsa var. rugosperma
Carex trichocarpa
Carex trisperma
Carex utriculata
Carex vulpinoidea
Carex woodii
Cyperus bipartitus
Cyperus esculentus
Dulichium arundinaceum
Eleocharis acicularis
Eleocharis intermedia
Eriophorum virginicum
Eriophorum viridicarinatum
Schoenoplectiella smithii var. smithii
Schoenoplectus tabernaemontani
Scirpus atrovirens
Scirpus cyperinus
Order
Family
Juniperus virginiana
Luzula acuminata
Luzula multiflora subsp. multiflora
Order
Family
Iris spp.
Iris versicolor
Sisyrinchium angustifolium
Sisyrinchium montanum
Sisyrinchium mucronatum
Family
Clintonia borealis
Erythronium albidum
Erythronium americanum subsp. americanum
Hemerocallis fulva
Hemerocallis lilioasphodelus
Lilium canadense
Lilium michiganense
Lilium philadelphicum
Lilium superbum
Medeola virginiana
Polygonatum biflorum
Polygonatum pubescens
Streptopus lanceolatus var. lanceolatus
Trillium erectum
Trillium grandiflorum
Trillium spp.
Uvularia grandiflora
Family
Anticlea elegans var. glaucus
Family
Smilax herbacea
Smilax tamnoides
Order
Family
Najas flexilis
Family
Potamogeton amplifolius
Potamogeton pusillus subsp. pusillus
Order
Family
Aplectrum hyemale
Epipactis helleborine
Galearis spectabilis
Liparis loeselii
Platanthera dilatata
Platanthera hyperborea
Platanthera nivea
Spiranthes lucida
Order
Family
Juncus acuminatus
Juncus articulatus
Juncus brachycarpus
Juncus bufonius
Juncus dudleyi
Juncus effusus
Juncus nodosus
Juncus tenuis
Family
Agrostis capillaris
Agrostis gigantea
Agrostis stolonifera
Alopecurus aequalis
Andropogon gerardii
Avena fatua
Avena sativa
Brachyelytrum erectum
Briza maxima
Bromus ciliatus
Bromus latiglumis
Bromus pubescens
Calamagrostis canadensis
Cinna arundinacea
Dactylis glomerata
Danthonia spicata
Dichanthelium dichotomum var. dichotomum
Dichanthelium latifolium
Dichanthelium linearifolium
Digitaria sanguinalis
Eleusine indica
Elymus canadensis
Elymus hystrix
Elymus lanceolatus subsp. psammophilus
Elymus repens
Elymus riparius
Elymus virginicus var. virginicus
Festuca rubra subsp. rubra
Glyceria borealis
Glyceria canadensis
Glyceria maxima
Glyceria septentrionalis
Glyceria striata
Leersia oryzoides
Lolium perenne
Milium effusum
Muhlenbergia glomerata
Muhlenbergia mexicana var. mexicana
Muhlenbergia tenuiflora
Oryzopsis asperifolia
Panicum capillare
Panicum virgatum
Patis racemosa
Phalaris arundinacea
Poa compressa
Poa nemoralis
Poa pratensis subsp. pratensis
Poa ssp.
Schizachne purpurascens
Schizachyrium scoparium
Setaria viridis
Sorghastrum nutans
Sporobolus vaginiflorus
Bromus inermis
Bromus kalmii
Echinochloa crus-galli
Phleum pratense
Phragmites australis
Setaria pumila
Spartina pectinata
Family
Typha angustifolia
Typha latifolia
Order
Family
Sparganium americanum
Sparganium eurycarpum
Class
Order
Family
Aegopodium podagraria
Angelica atropurpurea
Cicuta bulbifera
Cicuta maculata
Conioselinum chinense
Cryptotaenia canadensis
Daucus carota
Erigenia bulbosa
Heracleum mantegazzianum
Osmorhiza claytonii
Sanicula canadensis var. grandis
Sanicula marilandica
Sium suave
Thaspium trifoliatum var. aureum
Zizia aurea
Family
Aralia elata
Aralia nudicaulis
Aralia racemosa subsp. racemosa
Hydrocotyle americana
Panax quinquefolius
Panax trifolius
Order
Family
Asarum canadense
Order
Family
Achillea millefolium
Ageratina altissima
Ambrosia artemisiifolia
Ambrosia trifida
Antennaria neglecta
Arctium lappa
Arctium minus
Artemisia campestris subsp. caudata
Artemisia vulgaris
Aster spp.
Bidens cernua
Bidens connata
Bidens frondosa
Bidens ssp.
Bidens vulgata
Carduus nutans
Centaurea jacea
Centaurea stoebe subsp. micranthos
Cichorium intybus
Cirsium arvense
Cirsium discolor
Cirsium muticum
Cirsium oleraceum
Cirsium pumilum var. hilii
Cirsium vulgare
Echinacea purpurea
Erechtites hieraciifolius
Erigeron annuus
Erigeron canadensis
Erigeron philadelphicus var. philadelphicus
Erigeron pulchellus
Erigeron strigosus
Eupatorium hyssopifolium
Eupatorium perfoliatum
Eurybia macrophylla
Euthamia graminifolia
Eutrochium maculatum var. maculatum
Eutrochium purpureum var. purpureum
Helenium flexuosum
Helianthus annuus
Helianthus decapetalus
Helianthus giganteus
Helianthus tuberosus
Heliopsis helianthoides
Hieracium umbellatum
Hieracium vulgatum
Inula helenium
Lactuca canadensis
Lactuca serriola
Lapsana communis
Leucanthemum vulgare
Liatris cylindracea
Matricaria discoidea
Nabalus albus
Nabalus altissimus
Onopordum acanthium
Packera aurea
Pilosella ×floribunda
Pilosella aurantiaca
Pilosella caespitosa
Pilosella officinarum
Pilosella piloselloides subsp. piloselloides
Pilosella piloselloides subsp. praealta
Prenanthes spp.
Ratibida pinnata
Rudbeckia hirta
Rudbeckia laciniata
Solidago altissima var. altissima
Solidago caesia
Solidago canadensis
Solidago flexicaulis
Solidago gigantea
Solidago hispida
Solidago juncea
Solidago nemoralis subsp. nemoralis
Solidago patula
Solidago ptarmicoides
Solidago rigida
Solidago rugosa subsp. rugosa
Solidago spp.
Solidago ulmifolia
Sonchus arvensis subsp. arvensis
Sonchus asper
Sonchus oleraceus
Symphyotrichum cordifolium
Symphyotrichum ericoides var. ericoides
Symphyotrichum laeve
Symphyotrichum lanceolatum
Symphyotrichum lateriflorum var. lateriflorum
Symphyotrichum lateriflorum var. hirsuticaule
Symphyotrichum novae-angliae
Symphyotrichum novi-belgii
Symphyotrichum oolentangiense var. oolentangiense
Symphyotrichum oolentangiense
Symphyotrichum pilosum var. pilosum
Symphyotrichum puniceum
Symphyotrichum racemosum
Symphyotrichum spp.
Symphyotrichum urophyllum
Tanacetum vulgare
Taraxacum officinale
Tragopogon dubius
Tragopogon pratensis
Tussilago farfara
Xanthium strumarium
Family
Lobelia siphilitica
Order
Family
Hackelia virginiana
Myosotis scorpioides
Order
Family
Hesperis matronalis
Rorippa sylvestris
Order
Family
Campanula rapunculoides
Campanula rotundifolia
Lobelia cardinalis
Lobelia inflata
Order
Family
Alliaria petiolata
Barbarea vulgaris
Berteroa incana
Brassica juncea
Brassica rapa
Capsella bursa-pastoris
Cardamine bulbosa
Cardamine concatenata
Cardamine diphylla
Cardamine maxima
Cardamine pensylvanica
Erysimum cheiranthoides
Nasturtium officinale
Rorippa palustris subsp. palustris
Sinapis alba
Sinapis arvensis
Sisymbrium altissimum
Thlaspi arvense
Turritis glabra
Order
Family
Amaranthus albus
Amaranthus retroflexus
Amaranthus tuberculatus
Family
Arenaria serpyllifolia
Cerastium arvense subsp. arvense
Cerastium fontanum subsp. vulgare
Dianthus armeria
Dianthus barbatus
Saponaria officinalis
Silene latifolia
Silene vulgaris
Stellaria graminea
Family
Chenopodium album
Family
Phytolacca americana var. americana
Family
Persicaria hydropiper
Persicaria pensylvanica
Rumex britannica
Rumex crispus
Rumex obtusifolius
Rumex verticillatus
Family
Claytonia virginica
Order
Family
Ilex verticillata
Family
Celastrus scandens
Euonymus alatus
Euonymus atropurpureus
Euonymus obovatus
Order
Family
Cornus alternifolia
Cornus canadensis
Cornus obliqua
Cornus racemosa
Cornus rugosa
Cornus stolonifera
Order
Family
Echinocystis lobata
Order
Family
Viburnum acerifolium
Viburnum lantana
Viburnum lentago
Viburnum opulus
Viburnum opulus var. americanum
Viburnum rafinesquianum
Family
Caprifoliaceae spp.
Diervilla lonicera
Dipsacus sativus
Linnaea borealis subsp. longiflora
Lonicera canadensis
Lonicera dioica
Lonicera morrowii
Lonicera tatarica
Sambucus canadensis
Sambucus nigra
Sambucus racemosa subsp. pubens
Symphoricarpos albus
Symphoricarpos occidentalis
Triosteum aurantiacum
Family
Dipsacus fullonum
Family
Valeriana edulis subsp. cilata
Order
Family
Impatiens capensis
Family
Epigaea repens
Gaultheria hispidula
Gaultheria procumbens
Vaccinium angustifolium
Vaccinium myrtilloides
Family
Lysimachia arvensis
Family
Pyrola elliptica
Pyrola spp.
Order
Family
Euphorbia cyparissias
Euphorbia esula
Checklist of species observed or collected at the rare Charitable Research Reserve in Cambridge, Ontario, Canada. The second of two checklists for Kingdom Plantae, this checklist contains records from Phyla Magnoliophyta (Class Magnoliopsida), Marchantiophyta, Pinophyta, and Pteridophyta.
Phylum
Kingdom
Class
Order
Family
Amorpha canescens
Amphicarpaea bracteata
Apios americana
Desmodium canadense
Glycyrrhiza lepidota
Lespedeza capitata
Lotus corniculatus
Medicago lupulina
Medicago sativa subsp. sativa
Melilotus albus
Robinia pseudoacacia
Trifolium aureum
Trifolium campestre
Trifolium hybridum
Trifolium pratense
Trifolium repens
Vicia americana
Vicia cracca
Order
Family
Alnus glutinosa
Alnus incana subsp. rugosa
Betula alleghaniensis
Betula papyrifera
Betula pendula
Betula populifolia
Betula pumila
Carpinus caroliniana subsp. virginiana
Corylus americana
Corylus cornuta subsp. cornuta
Ostrya virginiana
Family
Fagus grandifolia
Quercus alba
Quercus bicolor
Quercus ellipsoidalis
Quercus macrocarpa
Quercus palustris
Quercus rubra
Quercus velutina
Family
Juglans nigra
Order
Family
Apocynum androsaemifolium
Apocynum cannabinum
Apocynum sp.
Vinca minor
Family
Asclepias exaltata
Asclepias incarnata subsp. incarnata
Asclepias syriaca
Asclepias tuberosa
Family
Gentiana andrewsii
Gentiana rubricaulis
Gentianella quinquefolia subsp. quinquefolia
Gentianopsis crinita
Gentianopsis virgata subsp. virgata
Family
Galium aparine
Galium asprellum
Order
Family
Impatiens glandulifera
Impatiens pallida
Family
Geranium maculatum
Geranium robertianum
Family
Oxalis corniculata
Oxalis montana
Oxalis stricta
Order
Family
Myriophyllum heterophyllum
Myriophyllum verticillatum
Order
Family
Platanus occidentalis
Order
Family
Carya cordiformis
Carya glabra
Carya ovata var. ovata
Juglans cinerea
Order
Family
Anchusa arvensis
Cynoglossum officinale
Echium vulgare
Lithospermum parviflorum
Myosotis laxa
Myosotis verna
Symphytum officinale
Family
Agastache nepetoides
Ajuga reptans
Blephilia hirsuta
Clinopodium vulgare
Collinsonia canadensis
Galeopsis tetrahit
Glechoma hederacea
Lamium amplexicaule
Leonurus cardiaca
Lycopus americanus
Lycopus asper
Lycopus uniflorus
Melissa officinalis
Mentha arvensis
Mentha canadensis
Mentha sp.
Mentha spicata
Monarda didyma
Monarda fistulosa
Nepeta cataria
Physostegia virginiana subsp. virginiana
Prunella vulgaris subsp. vulgaris
Prunella vulgaris subsp. lanceolata
Prunella vulgaris
Pycnanthemum virginianum
Scutellaria galericulata
Scutellaria lateriflora
Stachys hispida
Stachys palustris
Teucrium canadense subsp. canadense
Family
Ligustrum vulgare
Syringa vulgaris
Family
Linaria vulgaris
Plantago lanceolata
Plantago major
Plantago rugelii
Veronica anagallis-aquatica
Family
Verbascum phoeniceum
Verbascum thapsus
Veronica chamaedrys
Veronica officinalis
Veronica serpyllifolia
Family
Verbena hastata
Verbena simplex
Verbena stricta
Verbena urticifolia
Order
Family
Lindera benzoin
Order
Family
Linum virginianum
Order
Family
Asimina triloba
Order
Family
Hypericum ascyron
Hypericum mutilum subsp. mutilum
Hypericum perforatum
Hypericum punctatum
Family
Populus alba
Populus balsamifera
Populus deltoides
Populus grandidentata
Populus nigra
Populus tremuloides
Order
Family
Abutilon theophrasti
Malva moschata
Family
Tilia americana
Order
Family
Lythrum salicaria
Family
Chamerion angustifolium subsp. angustifolium
Circaea alpina
Circaea canadensis subsp. canadensis
Epilobium ciliatum subsp. glandulosum
Epilobium coloratum
Epilobium hirsutum
Epilobium leptophyllum
Epilobium strictum
Ludwigia polycarpa
Oenothera biennis
Oenothera serrulata
Family
Dirca palustris
Order
Family
Brasenia schreberi
Order
Family
Chelidonium majus
Dicentra canadensis
Dicentra cucullaria
Sanguinaria canadensis
Order
Family
Saururus cernuus
Order
Family
Callitriche palustris
Chelone glabra
Digitalis lanata
Digitalis lutea
Digitalis purpurea
Order
Family
Polygala spp
Polygala verticillata var. verticillata
Polygaloides paucifolia
Order
Family
Fallopia convolvulus
Fallopia scandens
Persicaria hydropiperoides
Persicaria maculosa
Persicaria punctata
Persicaria sagittata
Polygonum aviculare
Order
Family
Lysimachia borealis
Lysimachia ciliata
Lysimachia nummularia
Lysimachia thyrsiflora
Primula mistassinica
Order
Family
Berberis canadensis
Berberis thunbergii
Berberis vulgaris
Caulophyllum thalictroides
Podophyllum peltatum
Family
Menispermum canadense
Family
Actaea pachypoda
Actaea rubra
Actaea spp.
Anemone acutiloba
Anemone americana
Anemone canadensis
Anemone cylindrica
Anemone quinquefolia
Anemone virginiana var. virginiana
Anemone virginiana var. alba
Aquilegia canadensis
Caltha palustris
Clematis virginiana
Coptis trifolia
Ranunculus abortivus
Ranunculus acris
Ranunculus hispidus var. caricetorum
Ranunculus recurvatus var. recurvatus
Ranunculus repens
Ranunculus sceleratus var. sceleratus
Thalictrum dasycarpum
Thalictrum dioicum
Thalictrum pubescens
Thalictrum thalictroides
Order
Family
Elaeagnus angustifolia
Elaeagnus umbellata
Shepherdia canadensis
Family
Frangula alnus
Rhamnus alnifolia
Order
Family
Humulus lupulus
Family
Sedum acre
Family
Ribes americanum
Ribes cynosbati
Ribes glandulosum
Ribes hirtellum
Ribes lacustre
Ribes oxyacanthoides var. oxycanthoides
Ribes rubrum
Ribes ssp.
Ribes triste
Family
Philadelphus coronarius
Family
Rhamnus cathartica
Family
Agrimonia gryposepala
Amelanchier arborea
Amelanchier laevis
Amelanchier spicata
Aruncus dioicus
Crataegus chrysocarpa
Crataegus crus-galli
Crataegus dissona
Crataegus punctata
Crataegus spp.
Dasiphora fruticosa
Fragaria vesca
Fragaria virginiana
Geum aleppicum
Geum canadense
Geum fragarioides
Geum laciniatum
Geum rivale
Geum urbanum
Malus coronaria
Malus pumila
Physocarpus opulifolius
Potentilla anserina subsp. anserina
Potentilla argentea
Potentilla norvegica
Potentilla recta
Potentilla simplex
Prunus americana
Prunus avium
Prunus mahaleb
Prunus pensylvanica
Prunus serotina
Prunus virginiana subsp. virginiana
Pyrus communis
Rosa blanda
Rosa cinnamomea
Rosa multiflora
Rosa palustris
Rosa rugosa
Rosa virginiana
Rubus allegheniensis
Rubus canadensis
Rubus flagellaris
Rubus hispidus
Rubus idaeus
Rubus occidentalis
Rubus pubescens
Rubus repens
Sorbus americana
Sorbus aucuparia
Spiraea alba
Family
Micranthes virginiensis
Mitella diphylla
Mitella nuda
Parnassia glauca
Tiarella cordifolia
Order
Family
Galium circaezans
Galium mollugo
Galium palustre
Galium tinctorium
Galium trifidum subsp. trifidum
Galium triflorum
Mitchella repens
Order
Family
Salix ×fragilis
Salix alba
Salix amygdaloides
Salix bebbiana
Salix candida
Salix cordata
Salix discolor
Salix eriocephala
Salix exigua
Salix humilis var. humilis
Salix lucida
Salix nigra
Salix pedicellaris
Salix petiolaris
Salix purpurea
Salix serissima
Salix viminalis
Order
Family
Acer nigrum
Acer platanoides
Acer rubrum
Acer saccharinum
Acer saccharum
Acer spicatum
Family
Rhus aromatica
Rhus typhina
Toxicodendron radicans subsp. negundo
Toxicodendron radicans var. rydbergii
Toxicodendron vernix
Family
Zanthoxylum americanum
Family
Acer negundo
Aesculus hippocastanum
Family
Staphylea trifolia
Order
Family
Myriophyllum sibiricum
Order
Family
Fraxinus americana
Fraxinus nigra
Fraxinus pennsylvanica
Family
Conopholis americana
Epifagus virginiana
Family
Aureolaria flava
Aureolaria virginica
Pedicularis canadensis
Penstemon digitalis
Penstemon hirsutus
Scrophularia lanceolata
Scrophularia marilandica
Verbascum blattaria
Veronica americana
Order
Family
Calystegia spithamaea
Convolvulus arvensis
Cuscuta gronovii
Family
Hydrophyllum canadense
Hydrophyllum virginianum
Family
Phlox subulata subsp. subulata
Family
Physalis heterophylla
Solanum dulcamara
Solanum lycopersicum
Order
Family
Morus alba
Family
Celtis occidentalis
Ulmus americana
Ulmus pumila
Ulmus rubra
Ulmus thomasii
Family
Boehmeria cylindrica
Laportea canadensis
Pilea pumila
Urtica dioica subsp. gracilis
Urtica dioica subsp. dioica
Order
Family
Sicyos angulatus
Family
Viola adunca var. adunca
Viola canadensis
Viola cucullata
Viola labradorica
Viola macloskeyi
Viola nephrophylla
Viola pubescens
Viola renifolia
Viola rostrata
Viola sororia
Viola sororia var. affinis
Viola spp.
Order
Family
Parthenocissus quinquefolia
Vitis riparia
Order
Family
Phryma leptostachya
Order
Family
Hamamelis virginiana
Phylum
Class
Order
Family
Aneura pinguis
Order
Family
Lophocolea heterophylla
Family
Porella platyphylla
Family
Radula complanata
Order
Family
Frullania eboracensis
Class
Order
Family
Reboulia hemisphaerica
Family
Conocephalum conicum
Family
Marchantia polymorpha
Phylum
Class
Order
Family
Juniperus communis
Juniperus communis var. depressa
Order
Family
Thuja occidentalis
Family
Larix decidua
Larix laricina
Picea abies
Picea glauca
Picea pungens
Pinus banksiana
Pinus resinosa
Pinus strobus
Pinus sylvestris
Tsuga canadensis
Phylum
Class
Order
Family
Botrypus virginianus
Order
Family
Asplenium platyneuron
Asplenium trichomanes subsp. trichomanes
Asplenium viride
Family
Pteridium aquilinum var. latiusculum
Family
Athyrium filix-femina var. angustum
Cystopteris bulbifera
Cystopteris fragilis
Deparia acrostichoides
Dryopteris ×triploidea
Dryopteris carthusiana
Dryopteris clintoniana
Dryopteris cristata
Dryopteris goldiana
Dryopteris intermedia
Dryopteris marginalis
Gymnocarpium dryopteris
Matteuccia struthiopteris var. pensylvanica
Onoclea sensibilis
Polystichum acrostichoides
Family
Osmunda claytoniana
Osmunda regalis var. spectabilis
Family
Polypodium virginianum
Family
Adiantum pedatum
Pellaea glabella
Family
Thelypteris noveboracensis
Thelypteris palustris var. pubescens
Class
Order
Family
Osmundastrum cinnamomea
Osmundastrum cinnamomea
Analysis
The two surveying strategies – a four-month long terrestrial arthropod survey, followed by a concentrated bioblitz targeting a variety of taxa – resulted in 25,287 and 3,502 specimens barcoded, respectively, out of a total of 32,645 specimens collected. Observations of an additional 125 species and two higher taxa (for which no voucher specimen was kept) were recorded at the bioblitz (Suppl. material
The most diverse groups were Ichneumonidae, Chironomidae, and Cecidomyiidae with 188, 365, and 584 BINs respectively. In terms of abundance, the three groups Sciaridae, Cecidomyiidae, and Chironomidae had the largest number of specimens with 1,528, 2,477, and 9,636 specimens. The most abundant BINs were BOLD:ACC0651 (Thripidae: Taeniothrips inconsequens), BOLD:AAD5253 (Chironomidae: Thienemanniella xena), and BOLD:AAP5920 (Chironomidae: Cricotopus triannulatus) with 349, 636, and 1619 specimens collected. For these three BINs, and many similarly abundant BINs, the presence of closely allied and morphologically similar taxa makes oversampling of these exceptionally common species unavoidable.
Combining all existing data results in a final tally of 3,348 species (Table
Summary of species inventory for rare Charitable Research Reserve in Cambridge, Ontario, Canada, following the present study.
| Major Group | Common Name | No. on previous inventory | No. on 2015 surveys | New Species | New Total |
| Annelida | Earthworms | 0 | 2 | 2 |
2 |
| Arthropoda:Arachnida | Spiders and others | 0 | 198 | 198 | 198 |
| Arthropoda:Crustacea | Crustaceans | 0 | 7 | 7 | 7 |
| Arthropoda:Entognatha | Collembola | 0 | 9 | 9 | 9 |
| Arthropoda:Insecta | Insects |
832 |
895 | 778 | 1,610 |
| Arthropoda:Myriapoda | Millipedes, Centipedes | 0 | 6 | 6 | 6 |
| Chordata:Actinopterygii | Fishes | 28 | 14 | 3 | 31 |
| Chordata:Amphibia | Amphibians | 13 | 2 | 0 | 13 |
| Chordata:Aves | Birds | 231 | 87 | 0 | 231 |
| Chordata:Mammalia | Mammals | 37 | 6 | 1 | 38 |
| Chordata:Reptilia | Reptiles | 10 | 0 | 0 | 10 |
| Fungi | Fungus, Lichens | 191 | 84 | 60 | 251 |
| Mollusca | Snails, Clams | 0 | 18 | 18 | 18 |
| Plantae | Plants, Mosses, Liverworts | 901 | 103 | 20 | 921 |
| Protozoa | Protozoans | 3 | 3 | 0 | 3 |
| Total Species | 2,246 | 1,433 | 1,102 | 3,348 |
Discussion
The present study has conducted an expansive biotic survey, released the data in several public biodiversity data repositories, and published the unique process and findings – all within a relatively modest timeframe. This model for rapid generation and dissemination of critical biodiversity data could be followed to conduct regional assessments of biodiversity status and change, and potentially aid in evaluating progress towards the Aichi Targets of the Strategic Plan for Biodiversity 2011–2020. To fully appreciate this approach, a closer look at four elements is presented: highlights of the biotic inventory, the multiple levels of acceleration, future improvements to the resource, and the utility of the resource going forward.
Resource Highlights
The biotic inventory of rare Charitable Research Reserve performed between May and August 2015 was noteworthy for a number of reasons. The taxonomic scope of the surveys, which covered fourteen phyla over several kingdoms, was only made possible by the integration of DNA barcoding and by assembling a diverse group of experts. Taxa that appeared under-represented on the prior species inventory were targeted where possible, and in many cases, increased significantly. Spiders (Araneae) for instance were completely absent from the prior inventory. Using the expertise of single specialist, focused collecting efforts, and a comprehensive barcode reference library for Canadian spiders (
Rapid Creation of the Resource
The rapidness of conducting this inventory was due to the acceleration of several steps that comprise the procedure. Firstly, several types of passive traps were employed to acquire large sample sizes for minimal collector effort. For instance, malaise traps often collect 2,000 specimens in a single week with only minutes of servicing time. Secondly, as discussed above, the addition of DNA barcoding streamlines sorting of the material, dividing the specimens into distinct units, minimizing the total number of specimens that require examination. Thirdly, in addition to the initial sorting, barcoding also provides nearly instant taxonomic assignment by querying against reference libraries using BOLD; the resolution of the assignment for queried records depends on the completeness of the reference library. Fourthly, the processing of the bioblitz material and analyses of the DNA barcode data were accelerated at all possible stages to test how quickly a large volume of specimens could proceed through all steps. The generation of data following the bioblitz was impressive: tissue sampling and lysis within 12 hours; DNA extraction and PCR within 24 h; cycle sequencing, cleanup and sequencer loading by 48 h; edited and validated sequences on BOLD by 72 h; taxonomic assignment, data release and manuscript presubmission by 96 h; final manuscript submission to BDJ by 108 h. The laboratory steps are mostly accelerated by automation, while data sharing is greatly facilitated by data release platforms such as BOLD and the Integrated Publishing Toolkit of Canadensys (
Refinement of the Resource
Many BINs are presently identified only to family, subfamily, or at most, genus level. It's important to note that the inventoried taxa that lack species-level determination, including intractable groups such as mites and midges, will be refined over time. Not only does DNA barcoding, empowered by the BIN system, roughly sort the material to direct and minimize the efforts of taxonomic experts (
In addition to this passive approach to refining the taxonomy of these specimens, we will be actively pursuing expert determinations for the unnamed BINs. For many groups, the experts among our coauthors will be able to provide species level resolution in a short period of time (e.g., J.F.-T. specializes in Braconidae, particularly the subfamily Microgastrinae). For other BINs, and over a longer period of time, it will be necessary to solicit determinations from our network of collaborators that specialize on Nearctic taxa. The public release of these data to multiple data repositories should also aid in recruiting active specialists presently outside our network. The vouchered material, all deposited in a single repository (BIOUG), can be quickly assembled and loaned out, along with the associated data and files that may assist in determinations (e.g., tree files and image libraries: Suppl. materials
When a significant number of species and other taxon categories become available, the refined dataset can be updated in the various data repositories and potentially in a second, updated version of the paper in BDJ. Any subsequent versions – each with a separate Digital Object Identifier (DOI) – would be linked to the original paper, allowing for continuing improvement and additions to the species list for the reserve. It is important to emphasize that the taxa awaiting species-level determination still have persistent identifiers as part of the BIN system; each BIN is assigned a BOLD-generated uniform resource identifier (URI) upon its establishment (e.g. BOLD:AAA0001). Until the species binomial is determined or described, this URI can be used in its place. For example, this URI permits assessing and comparing local diversity (
Utility of the Resource
While the overarching objective of the study was to develop, assess and demonstrate a model for conducting and disseminating DNA barcode-assisted biotic surveys, a valuable resource was created in the process. The rare Charitable Research Reserve provides a unique urban reserve with the infrastructure necessary to conduct research in its diversity of habitats; a barcode reference library and updated species inventory can now both be added to the infrastructure shared with their researchers and educators. Ecological studies in particular, such as the ongoing prairie community field experiments (e.g.,
Acknowledgements
Financial support was provided by the Ontario Ministry of Research and Innovation and by the government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life project. Anne McCain Evans and Chris Evans provided support for the BIObus program (www.biobus.ca), whose students conducted much of the field work. A biotic survey and bioblitz of this magnitude could not be completed without the help of many volunteers; we would like to thank the following for their contributions to this study: Reza Zahiri, Carleigh Pope, Cheyanne Richardson, Christine Thompson, Dan Radoslav, Dave Achtymichuk, Erika Kastner, Kim Robichaud, Mike Achtymichuk, Tim Skuse, Lisandra Gal, and Suz Bateson. We would also like to thank our reviewers for their helpful suggestions and contributions to this manuscript: Jeremy Miller, Pavel Stoev, and Torsten Dikow.
Author contributions
Author contributions: P.D.N.H. and J.R.D. designed the study; J.S., C.N.S. and M.R.Y. oversaw specimen and sequence analyses; A.C.T., J.R.D. and V.L.-B. conducted the analyses. J.M. and E.S. provided specimen images. All authors discussed the results and contributed to specimen collection, identifications, and to the manuscript, which was written by J.R.D., R.D., J.F.-T.
References
- International Barcode of Life: Evolution of a global research community.Genome58:151‑162. https://doi.org/10.1139/gen-2015-0094
- Untangling taxonomy: a DNA barcode reference library for Canadian spiders.Molecular Ecology Resourcesn/a:n/a‑n/a. https://doi.org/10.1111/1755-0998.12444
- Error Cascades in the Biological Sciences: The Unwanted Consequences of Using Bad Taxonomy in Ecology.AMBIO: A Journal of the Human Environment37(2):114‑118. https://doi.org/10.1579/0044-7447(2008)37[114:ecitbs]2.0.co;2
- The adventive genus Xantholinus Dejean (Coleoptera, Staphylinidae, Staphylininae in North America: new records and a synthesis of distributional data.ZooKeys65:51‑61. https://doi.org/10.3897/zookeys.65.574
- Treasure island: pinning down a model ecosystem.Nature439(7075):378‑379. https://doi.org/10.1038/439378a
- EstimateS turns 20: statistical estimation of species richness and shared species from samples, with non-parametric extrapolation.Ecography37(6):609‑613. https://doi.org/10.1111/ecog.00814
- From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity.Trends in Ecology & Evolution29(10):566‑571. https://doi.org/10.1016/j.tree.2014.08.001
- deWaard J, Ivanova N, Hajibabaei M, Hebert PN (2008) Assembling DNA Barcodes. Methods in Molecular Biology. https://doi.org/10.1007/978-1-59745-548-0_15
- In the dark in a large urban park: DNA barcodes illuminate cryptic and introduced moth species.Biodiversity and Conservation18(14):3825‑3839. https://doi.org/10.1007/s10531-009-9682-7
- A Rosetta Stone for Nature’s Benefits to People.PLOS Biology13(1):e1002040. https://doi.org/10.1371/journal.pbio.1002040
- Fazekas A, Kuzmina M, Newmaster S, Hollingsworth P (2012) DNA Barcoding Methods for Land Plants. Methods in Molecular Biology™. https://doi.org/10.1007/978-1-61779-591-6_11
- A Catalogue of the Cecidomyiidae (Diptera) of the world. 3rd edition.Digital version 2.
- Trophic island biogeography drives spatial divergence of community establishment.Ecology95(10):2870‑2878. https://doi.org/10.1890/13-1683.1
- Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences270(1512):313‑321. https://doi.org/10.1098/rspb.2002.2218
- Genome size evolution in Ontario ferns (Polypodiidae): evolutionary correlations with cell size, spore size, and habitat type and an absence of genome downsizing.Genome57(10):555‑566. https://doi.org/10.1139/gen-2014-0090
- Refining the DNA barcode for land plants.Proceedings of the National Academy of Sciences108(49):19451‑19452. https://doi.org/10.1073/pnas.1116812108
- A DNA barcode for land plants.Proceedings of the National Academy of Sciences106(31):12794‑12797. https://doi.org/10.1073/pnas.0905845106
- Pre-made frozen PCR and sequencing plates.http://www.dnabarcoding.ca/CCDB_DOCS/CCDB_Advances_Methods_Release_No4_Dec1st_2006.pdf. Accessed on: 2015-8-17.
- An inexpensive, automation-friendly protocol for recovering high-quality DNA.Molecular Ecology Notes6(4):998‑1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
- Semi-automated, Membrane-Based Protocol for DNA Isolation from Plants.Plant Molecular Biology Reporter26(3):186‑198. https://doi.org/10.1007/s11105-008-0029-4
- CCDB Protocols. Glass Fiber Plate DNA Extraction Protocol For Plants, Fungi, Echinoderms and Mollusks, Manual Protocol Employing Centrifugation. http://www.ccdb.ca/CCDB_DOCS/CCDB_DNA_Extraction-Plants.pdf. Accessed on: 2015-8-17.
- Janzen D (1993) Taxonomy: Universal and Essential Infrastructure for Development and Management of Tropical Wildland Biodiversity. In: Sandlund OT, Schei PJ (Eds) Proc. Norway UNEP Expert Conference on Biodiversity, Trondheim.
- Setting up tropical biodiversity for conservation through non-damaging use: participation by parataxonomists.Journal of Applied Ecology41(1):181‑187. https://doi.org/10.1111/j.1365-2664.2004.00879.x
- Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity.Molecular Ecology Resources9:1‑26. https://doi.org/10.1111/j.1755-0998.2009.02628.x
- Janzen DH, Hallwachs W (1994) All Taxa Biodiversity Inventory (ATBI) of Terrestrial Systems: A Generic Protocol for Preparing Wildland Biodiversity for Non-Damaging Use. .
Report of a National Science Foundation Workshop, 16–18 April 1993, Philadelphia, Pennsylvania. On-line at www.all-species.org/content/reference/ATBI_Fin_Rep_8feb94_.pdf
- Ecology in the age of DNA barcoding: the resource, the promise and the challenges ahead.Molecular Ecology Resources14(2):221‑232. https://doi.org/10.1111/1755-0998.12173
- DNA barcodes for ecology, evolution, and conservation.Trends in Ecology & Evolution30(1):25‑35. https://doi.org/10.1016/j.tree.2014.10.008
- PCR Amplification for Plants and Fungi. http://www.ccdb.ca/CCDB_DOCS/CCDB_Amplification-Plants.pdf. Accessed on: 2015-8-17.
- Biodiversity inventories, indicator taxa and effects of habitat modification in tropical forest.Nature391(6662):72‑76. https://doi.org/10.1038/34166
- BioBlitz: Getting into Backyard Biodiversity.BioScience53(4):329. https://doi.org/10.1641/0006-3568(2003)053[0329:bgibb]2.0.co;2
- Measuring biological diversity.Blackwell Science,264pp.
- Evolution of endemism on a young tropical mountain.Nature000:1‑3. https://doi.org/10.1038/nature14949
- Canada's biodiversity: the variety of life, its status, economic benefits, conservation costs and unmet needs /.Canadian Museum of Nature,293pp. https://doi.org/10.5962/bhl.title.101447
- Low beta diversity of herbivorous insects in tropical forests.Nature448:692‑695.
- DNA barcoding and the mediocrity of morphology.Molecular Ecology Resources9:42‑50. https://doi.org/10.1111/j.1755-0998.2009.02631.x
- The Future of Biodiversity.Science269(5222):347‑350. https://doi.org/10.1126/science.269.5222.347
- The biodiversity of species and their rates of extinction, distribution, and protection.Science344(6187):1246752‑1246752. https://doi.org/10.1126/science.1246752
- Coupling non-destructive DNA extraction and voucher retrieval for small soft-bodied Arthropods in a high-throughput context: the example of Collembola.Molecular Ecology Resources10(6):942‑945. https://doi.org/10.1111/j.1755-0998.2010.2839.x
- BOLD: The Barcode of Life Data System (http://www.barcodinglife.org).Molecular Ecology Notes7(3):355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System.PLoS ONE8(7):e66213. https://doi.org/10.1371/journal.pone.0066213
- The GBIF Integrated Publishing Toolkit: Facilitating the Efficient Publishing of Biodiversity Data on the Internet.PLoS ONE9(8):e102623. https://doi.org/10.1371/journal.pone.0102623
- Specimens as primary data: museums and ‘open science’.Trends in Ecology & Evolution30(5):237‑238. https://doi.org/10.1016/j.tree.2015.03.002
- Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.Proceedings of the National Academy of Sciences of the United Sates of America109(16):6241‑6246. https://doi.org/10.1073/pnas.1117018109
- Lepidoptera of Great Smoky Mountains National Park: Methods and Results of the Inventory.Southeastern Naturalist6:193‑206. https://doi.org/10.1656/1528-7092(2007)6[193:logsmn]2.0.co;2
- The All Taxa Biological Inventory of the Great Smoky Mountains National Park.The Florida Entomologist84(4):556. https://doi.org/10.2307/3496388
- Lawton, J. H. and May, R. M. (Eds.). Extinction Rates. 1995. Oxford University Press, Oxford. xii + 233 pp. ISBN: 0-19-854829. X. Price: f17.95.Journal of Evolutionary Biology9(1):124‑126. https://doi.org/10.1046/j.1420-9101.1996.t01-1-9010124.x
- Exposing the structure of an Arctic food web.Ecology and EvolutionX:In press.
- Development of a Pollination Service Measurement (PSM) method using potted plant phytometry.Environmental Monitoring and Assessment186(8):5041‑5057. https://doi.org/10.1007/s10661-014-3758-x
- Revealing the Hyperdiverse Mite Fauna of Subarctic Canada through DNA Barcoding.PLoS ONE7(11):e48755. https://doi.org/10.1371/journal.pone.0048755
- Towards a comprehensive barcode library for arctic life - Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada.Frontiers in Zoology6(1):30. https://doi.org/10.1186/1742-9994-6-30
- Accelerated construction of a regional DNA-barcode reference library: caddisflies (Trichoptera) in the Great Smoky Mountains National Park.Journal of the North American Benthological Society30(1):131‑162. https://doi.org/10.1899/10-010.1
Supplementary materials
Species inventory for rare Charitable Research Reserve in Cambridge, Canada as of May 2015, prior to the present study.
Protocols for the bat component of rare BioBlitz
Details for laboratory work.
Summary data for the 28,789 specimens collected for the 2015 inventory, including BIN assignments and locality
Darwin core archive of 2015 rare species inventory
A list of all contributors who took part in the collection and identification of specimens collected as part of the 2015 inventory.
Human observations during rare BioBlitz, of mostly plants, birds, etc.
A neighbour-joining tree constructed from a single representative of each distinct BIN collected in rare, along with a single representative of clusters without BINs.
A collection of representative images of each BIN collected from the rare Charitable Research Reserve. BINs are listed in the same order as the tree. Specimens without BINs are not included.
The final combined inventory for rare Charitable Research Reserve as of August 2015; including previous checklist and all 2015 species added.
Lot and BIN data was downloaded for each specimen that received a BIN from BOLD Systems. It was formatted for input into EstimateS (Version 9.1.0) for creation of an accumulation curve.
Images of sampling sites, sampling techniques, specimen processing, and three new provincial spider reccords.