|
Biodiversity Data Journal :
Taxonomic Paper
|
|
Corresponding author: Ji-Chuan Kang (jckang@gzu.edu.cn)
Academic editor: Renan Barbosa
Received: 02 Feb 2021 | Accepted: 07 Mar 2021 | Published: 26 Mar 2021
© 2021 Lakmali Dissanayake, Nalin Wijayawardene, Monika Dayarathne, Milan Samarakoon, Dong-Qin Dai, Kevin Hyde, Ji-Chuan Kang
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Dissanayake LS, Wijayawardene NN, Dayarathne MC, Samarakoon MC, Dai D-Q, Hyde KD, Kang J-C (2021) Paraeutypella guizhouensis gen. et sp. nov. and Diatrypella longiasca sp. nov. (Diatrypaceae) from China. Biodiversity Data Journal 9: e63864. https://doi.org/10.3897/BDJ.9.e63864
|
|
Abstract
Background
In this study, we introduce a novel genus, Paraeutypella, of the family Diatrypaceae comprising three species viz. Paraeutypella guizhouensis sp. nov. and P. citricola (basionym: Eutypella citricola) and P. vitis (basionym: Sphaeria vitis). Diatrypella longiasca sp. nov. is also introduced, which forms a distinct clade in Diatrypella sensu stricto. The discovery of this new genus will contribute to expanding the knowledge and taxonomic framework of Diatrypaceae (Xylariales).
New information
Generic delimitations in Diatrypaceae are unsettled because the phylogeny has yet to be resolved using extensive taxon sampling and sequencing of ex-type cultures. During an investigation of xylarialean fungi, we collected eutypella-like fungi which is distinct from Eutypella sensu stricto in our phylogenetic analyses (ITS and β-tubulin), thus, introduced as Paraeutypella guizhouensis gen. et sp. nov.. Paraeutypella is characterised by having 4–25 perithecia in a stroma each with 3–6 sulcate, long ostiolar necks. Paraeutypella citricola comb. nov. (basionym: Eutypella citricola) is introduced on Acer sp. from China. Diatrypella longiasca sp. nov. is introduced as a new species in Diatrypella sensu stricto. which has 2–5 ascomata per stroma and long ascospores, unusual when compared to other Diatrypella species and distinct phylogenetically.
Keywords
Acer, morphology, novel taxa, phylogeny, Xylariales
Introduction
Diatrypaceae Nitschke (Ascomycota, Xylariales) comprises 21 genera and more than 1,500 species (
Members of Diatrypaceae are saprobes, pathogens or endophytes, associated with a wide range of hosts in terrestrial and aquatic environments (
Diatrypella was introduced by
In this study, we introduce a new genus, Paraeutypella, which shows eutypella-like morphology, but is distinct phylogenetically. Paraeutypella comprises three species viz. Paraeutypella guizhouensis sp. nov. and P. citricola (basionym: Eutypella citricola) and P. vitis (basionym: Sphaeria vitis). Diatrypella longiasca sp. nov. is also introduced, which forms a distinct clade in Diatrypella sensu stricto. Species novelties are confirmed by morphological comparisons along with micro-photographs and the phylogeny of combined ITS and β-tubulin sequence data.
Materials and methods
Sample collection and morphological observations
Dead twigs of Acer palmatum and undetermined plants were collected from China (Guiyang, Guizhou Province) during September to October 2019. Samples were observed with a stereomicroscope (SZX16, Olympus). Hand sections of the ascomata were mounted in distilled water and the following characters were measured: diameter and height of ascomata, width of the peridium, diameter and height of ostiolar necks, length and width of asci and ascospores. Melzer’s Reagent was used for testing the ascal apical ring reaction. Images were captured with a Canon EOS70D digital camera fitted to a compound microscope. Measurements were made with the Tarosoft (R) Image Frame Work programme and images used for figures processed with Adobe Photoshop CS6 software (Adobe Systems, USA). Single spore isolation was performed according to
DNA extraction, PCR amplifications and sequencing
Fungal isolates were grown on PDA for 3–4 weeks at 25°C and total genomic DNA was extracted from 50 to 100 mg of axenic mycelium scraped from the edges of the growing cultures (
DNA sequence data were obtained from the internal transcribed spacer (ITS) and partial β-tubulin gene. ITS and β-tubulin were amplified by using the primers ITS5/ITS4 (
Molecular phylogenetic analyses
Sequence alignment
The sequence data generated in this study were analysed with closely-related taxa retrieved from GenBank (Table
Taxa used in the phylogenetic analysis and their corresponding GenBank accession numbers.
|
Species |
Strain no. |
GenBank Accession no. |
Reference |
|
|
ITS |
β-tubulin |
|||
|
Allocryptovalsa cryptovalsoidea T |
HVFIG02 |
|
||
|
A. elaeidis |
MFLUCC 15-0707 |
|
||
|
Allodiatrype arengae T |
MFLUCC 15-0713 |
|
||
|
A. elaeidicola T |
MFLUCC 15-0737 |
|
||
|
A. elaeidis |
MFLUCC 15-0708b |
NA |
|
|
|
Anthostoma decipiens |
IPV-FW349 |
|
||
|
A. decipiens |
JL567 |
|
||
|
Cryptosphaeria eunomia |
CBS 216.87 |
NA |
|
|
|
C. var. eunomia |
CBS 223.87 |
NA |
|
|
|
Cryptovalsa ampelina |
A001 |
|
||
|
C. ampelina |
KHJ20 |
|
||
|
Diatrypasimilis australiensis T |
ATCC MYA 3540 |
NA |
|
|
|
Diatrype bullata |
UCDDCh400 |
|
||
|
D. disciformis T |
MFLUCC 15-0538 |
NA |
|
|
|
D. lijiangensis T |
MFLU 19-0717 |
|
||
|
D. stigma |
DCASH200 |
|
||
|
Diatrypella atlantica T |
HUEFS 194228 |
|
||
|
D. atlantica |
HUEFS 192148 |
|
||
|
D. delonicis T |
MFLUCC 15-1014 |
|
||
|
D. delonicis |
MFLU 16-1032 |
|
||
|
D. elaeidis T |
MFLUCC 15-0279 |
|
||
|
D. favacea |
ANM 96 |
NA |
|
|
|
D. frostii |
UFMGCB 1917 |
NA |
|
|
|
D. heveae T |
MFLUCC 17-0368 |
|
||
|
D. heveae |
MFLUCC 15-0274 |
|
||
|
D. iranensis T |
IRAN 2280C |
|
||
|
D. longiasca T |
KUMCC 20-0021 |
This study |
||
|
D. longiasca |
KUMCC 20-0022 |
This study |
||
|
D. macrospora T |
IRAN 2344C |
|
||
|
D. major |
ANM 1947 |
NA |
|
|
|
D. pulvinata |
H048 |
|
||
|
D. verruciformis |
UCROK1467 |
|
||
|
D. verruciformis |
UCROK754 |
|
||
|
D. vulgaris |
HVFRA02 |
|
||
|
D. vulgaris T |
HVGRF03 |
|
||
|
Eutypa laevata |
CBS 291.87 |
NA |
|
|
|
E. lata |
ATCC 28120 |
|
||
|
E. lata |
EP18 |
|
||
|
E. lata |
RGA01 |
|
||
|
E. lata var. aceri |
CBS 290.87 |
|
||
|
Eutypella caricae |
EL51C |
NA |
|
|
|
E. cerviculata |
EL59C |
NA |
|
|
|
E. cerviculata |
M68 |
NA |
|
|
|
E. leprosa |
EL54C |
NA |
|
|
|
E. leprosa |
ANM 85 |
NA |
|
|
|
E. microtheca |
ADEL200 |
|
||
|
E. microtheca |
BCMX01 |
|
||
|
E. parasitica |
CBS 210.39 |
NA |
|
|
|
E. persica T |
IRAN 2540C |
|
||
|
E. quercina T |
IRAN 2543C |
|
||
|
E. semicircularis T |
MP4669 |
NA |
|
|
|
E. tamaricis |
MFLUCC 14-0445 |
NA |
|
|
|
E. virescens |
CBS 205.36 |
|
||
|
Halocryptovalsa salicorniae |
MFLUCC 15-0185 |
|
||
|
Halodiatrype avicenniae |
MFLUCC 15-0948 |
|
||
|
H. salinicola T |
MFLUCC 15-1277 |
|
||
|
H. salinicola |
MFLUCC17-2468 |
NA |
|
|
|
Kretzschmaria deusta T |
CBS 826.72 |
|
||
|
Monosporascus cannonballus T |
ATCC 26931 |
NA |
Unpublished | |
|
M. cannonballus |
CMM3646 |
NA |
|
|
|
Neoeutypella baoshanensis |
MFLUCC 16-1002 |
NA |
|
|
|
N. baoshanensis T |
LC 12111 |
|
||
|
Paraeutypella citricolca |
HVGRF01 |
|
||
|
P. citricola |
HVVIT07 |
|
||
|
P. citricola |
IRAN 2340C |
|
||
|
P. citricola |
KUMCC 20-0023 |
This study |
||
|
P. citricola |
KUMCC 20-0024 |
This study |
||
|
P. guizhouensis T |
KUMCC 20-0016 |
This study |
||
|
P. guizhouensis |
KUMCC 20-0017 |
This study |
||
|
P. vitis |
UCD2291AR |
|
||
|
P. vitis |
UCD2428TX |
|
||
|
Pedumispora rhizophorae |
NA |
|
||
|
P. rhizophorae |
NA |
|
||
|
Peroneutypa curvispora |
HUEFS 136877 |
NA |
|
|
|
P. rubiformis T |
MFLUCC 17-2142 |
NA |
|
|
|
P. scoparia T |
MFLUCC 11-0478 |
NA |
|
|
|
Quaternaria quaternata |
CBS 278.87 |
NA |
|
|
|
Q. quaternata |
GNF13 |
|
||
| Xylaria hypoxylon T | CBS-122620 | KY610407 | KX271279 |
|
T: Types strains, newly-generated sequences are indicated in bold, NA: No sequence available in GenBank, ATCC: American Type Culture Collection, Manassas, USA, BCC: BIOTEC Culture Collection, Bangkok, Thailand, CBS: Centra albureau voor Schimmel cultures, Utrecht, The Netherlands, MFLU: Mae Fah Luang University, Chiang Rai, Thailand, CCMB: Bahia Culture Collection of Microorganisms, CMM: Culture Collection of Phytopathogenic Fungi “Prof. Maria Menezes,” Federal Rural University of Pernambuco, Brazil, MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand, HKAS: The Herbarium Mycologium, Institute of Microbiology Chinese Academy of Sciences, Beijing, China, HUEFS: Herbarium of the State University of Feira de Santana, IRAN: Iranian Fungal Culture Collection, Iranian Research Institute of Plant Protection, Tehran, Iran, KUMCC: Kunming Institute of Botany Culture Collection, Kunming, China.
Phylogenetic Analyses
Maximum Likelihood (ML) analysis was performed using RAxML-HPC2 on XSEDE (8.2.8) (
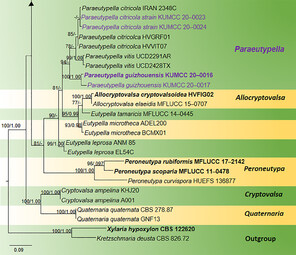
Phylogram generated from Maximum Likelihood (RAxML) analysis, based on ITS- β-tubulin matrix. ML bootstrap supports (≥ 70%) and Bayesian posterior probability (≥ 0.95) are indicated as ML/BYPP. The tree is rooted to Kretzschmaria deusta (CBS 826.72) and Xylaria hypoxylon (CBS 122620). Newly-generated strains are in red and type strains are in bold. The asterisks represent unstable species.
Taxon treatments
Diatrypella
Type species
Description
Notes – Diatrypella was introduced by Cesati & De Notaris (1863) and is typified as Diatrypella verruciformis (Ehrh.) Nitschke. There are 146 epithets listed in Index Fungorum (2020). This genus was established to accommodate members of stromatic Sphaeriales which were characterised by ovoid and numerous ascospores and we introduce a new species viz. Diatrypella longiasca, based on new collections from China.
Diatrypella longiasca , sp. nov.
-
kingdom: Fungi; phylum:Ascomycota; class:Sordariomycetes; order:Xylariales; family:Diatrypaceae; taxonRank:species; genus:Diatrypella; specificEpithet:longiasca; scientificNameAuthorship:L.S. Dissan., J.C. Kang & K.D. Hyde, sp. nov.; country:China; stateProvince:Guizhou Province; county:Guiyang; locality:Guizhou University Garden (South); identifiedBy:L.S. Dissanayake; institutionID:HMAS 290656; collectionID:HMAS 290658; institutionCode:Chinese Academy of Science, Kunming and Chinese Academy of Science Herbarium; collectionCode:Kunming Institute of Botany Culture Collection; datasetName:CLD 42
-
type: isotype; institutionID:HMAS 290658; collectionID:KUMCC 20-0022; institutionCode:Chinese Academy of Science, Kunming and Chinese Academy of Science Herbarium; collectionCode:Kunming Institute of Botany Culture Collection
Description
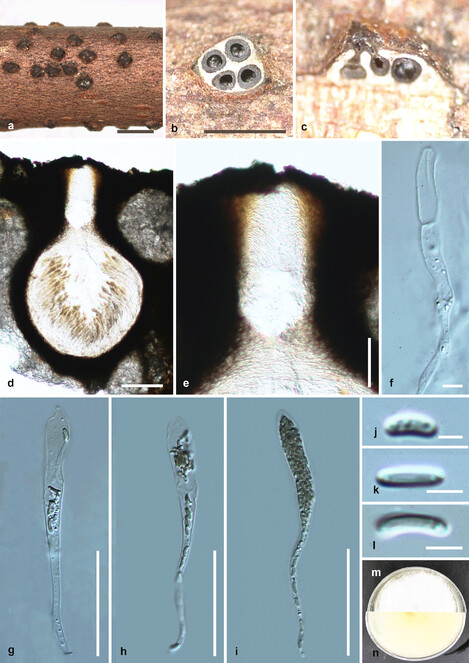
Saprobic on dead twigs (Fig.
Diatrypella longiasca (HMAS 290656, holotype) a. stromata on substrate; b. cross section of a stroma; c, d. vertical section through stroma showing ostiole and perithecia; e. ostiolar canal; f. paraphyses; g–i. asci; j–l. ascospores; m, n. culture on PDA from m above, n below after 6 weeks. Scale bars: 500 µm (a, b), 100 µm (d), 50 µm (e, g–i), 5 µm (f, j–l).
Culture characteristics – Colonies on PDA reaching 21 mm diam. after 2 weeks at 20–25oC, medium dense, circular to slightly irregular, slightly raised, cottony surface; colony from above: at first white, becoming buff; from below: yellowish white at margin, yellow to brown at centre; mycelium yellowish.
Etymology
The specific epithet longiasca refers to the long asci.
Notes
Diatrypella longiasca shares similar characters with D. vulgaris in having erumpent stromata through the bark often surrounded by remaining adherent epidermis or woody fragments and asci with many ascospores. However, D. vulgaris is different from D. longiasca in having longer ascospores (8–10 × 2–2.5 μm vs. 4–8 × 1–2 μm) (
Paraeutypella , gen. nov.
Type species
Description
Saprobic on dead twigs. Sexual morph: Stromata immersed in bark of dead branches, erumpent, solitary or aggregated. Ascomata with groups of 4–25 perithecia arranged in a valsoid configuration, surrounded by white, powdery entostroma, perithecial, black or brown, subglobose, clustered, immersed in stromata. Necks papillate, with an elongated ostiolar neck, central ostiolar canal filled with periphyses, 3–6 sulcate. Peridium composed of two layers of cells of textura angularis; inner layers cells hyaline or light brown, outer layers cells dark brown to black. Hamathecium composed of paraphyses arising from the base of perithecia, elongate, filiform, narrow, unbranched, septate, guttulate, narrowing and tapering towards apex. Asci 8-spored, unitunicate, thin-walled, clavate to cylindrical clavate or spindle-shaped, long pedicellate, apical rings J-. Ascospores overlapping biseriate, allantoid, slightly to moderately curved, allantoid, several oil droplets in each end, hyaline to light brown, sometimes yellow in mass, aseptate. Asexual morph: Coelomycetous. Conidiomata black, subconic, multiloculate, largely prosenchymatous, producing yellowish conidial masses. Conidiophores not recorded. Conidiogenous cells cylindrical, tapering, arising from pseudoparenchyma or interwoven hyphae, proliferating percurrently or sympodially, rarely both ways. Conidia hyaline, single-celled, slightly to moderately curved, with flattened bases, becoming guttulate (description of asexual morph adapted from
Etymology
With reference to the morphological resemblance to Eutypella.
Notes
Paraeutypella is introduced to accommodate three species viz. P. guizhouensis sp. nov., as well as P. citricola and P. vitis, two species previously placed in Eutypella sensu lato. Paraeutypella is typified by P. guizhouensis, which was collected from undetermined dead twigs. Paraeutypella can be distinguished from Eutypella species by stromata with perithecia in groups of 4–25 arranged in a valsoid configuration, 3–6 sulcate, long ostiolar necks, while stromata of Eutypella comprise groups of 20–70 perithecia having comparatively shorter ostiolar necks with sulcate or smooth ostiolar necks. Strains of both genera appear in distinct clades in a phylogeny based on ITS and Beta tubulin data (Fig.
Paraeutypella guizhouensis , sp. nov.
-
kingdom: Fungi; phylum:Ascomycota; class:Sordariomycetes; order:Xylariales; family:Diatrypaceae; genus:Paraeutypella; specificEpithet:guizhouensis; country:China; stateProvince:Guizhou Province; county:Guiyang; locality:Guizhou University Garden (North); habitat:Saprobic on dead twigs.; fieldNumber:CLD018; identifiedBy:L.S.Dissanayake; type:Holotype; institutionID:HMAS 290654; collectionID:KUMCC 20–0016; institutionCode:Chinese Academy of Science, Kunming and Chinese Academy of Science Herbarium; collectionCode:Kunming Institute of Botany Culture Collection; datasetName:CLD018
-
type: isotype; institutionID:HKAS 290655; collectionID:KUMCC 20-0017; institutionCode:Chinese Academy of Science, Kunming and Chinese Academy of Science Herbarium; collectionCode:Kunming Institute of Botany Culture Collection
Description
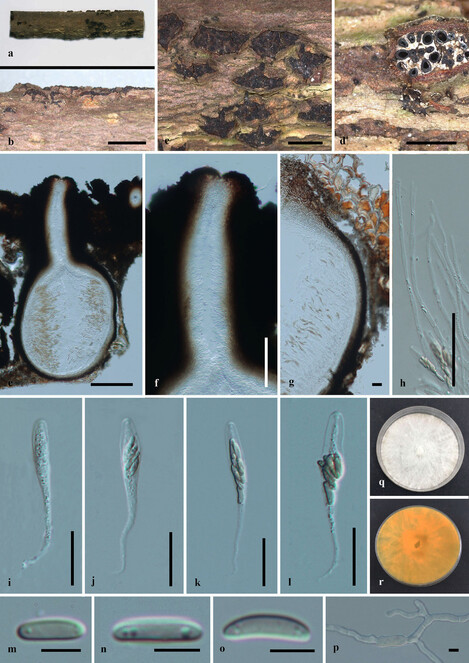
Saprobic on dead twigs (Fig.
Paraeutypella guizhouensis (HMAS 290654, holotype) a–c. stromata on substrate; d. cross section of a stromata; e. vertical section through an ascostroma showing ostioles and perithecia; f. ostiolar canal; g. peridium; h. paraphyses; i–l. asci; m–o. ascospores; p. germinating ascospore; q, r. cultures on PDA from above and below after 6 weeks. Scale bars: 500 µm (b–d), 200 µm (e), 100 µm (f), 20 µm (g–l), 5 µm (m–p).
Culture characteristics – Colonies on PDA, reaching 21 mm diam. after 2 weeks at 20–25oC, medium dense, circular to slightly irregular, slightly raised, cottony surface; colony from above: at first white, becoming buff; from below: yellowish-white at margin, yellow to brown at centre; mycelium yellowish.
Etymology
The specific epithet guizhouensis refers to the locality in which the fungus was collected.
Notes
Paraeutypella guizhouensis resembles P. vitis, which comprises stromata that are erumpent through bark, with elongated perithecial necks and allantoid, slightly to moderately curved ascospores (
Paraeutypella citricola () , comb. nov. ≡ Eutypella citricola in Anales del Museo Nacional de Buenos Aires 6: 245, (1898)
Nomenclature
= Eutypella citricola Syd. & P. Syd., Hedwigia 49: 80 (1909), nom. illegit., Art. 53.1
-
institutionID:
LPS-2120
-
kingdom: Fungi; phylum:Ascomycota; class:Sordariomycetes; order:Xylariales; family:Diatrypaceae; genus:Paraeutypella; specificEpithet:citricola; country:China; county:Guiyang; locationAccordingTo:Guizhou University Garden (South); year:2019; month:October; day:5; habitat:on a dead branch of Acer sp.; recordedBy:Nalin N. Wijayawardene; identifiedBy:L.S.Dissanayake; type:paratype; institutionID:HMAS 290660, HMAS 290659; collectionID:culture KUMCC 20–0024, KUMCC 20–0023; institutionCode:Chinese Academy of Science, Kunming and Chinese Academy of Science Herbarium; collectionCode:Kunming Institute of Botany Culture Collection
Description
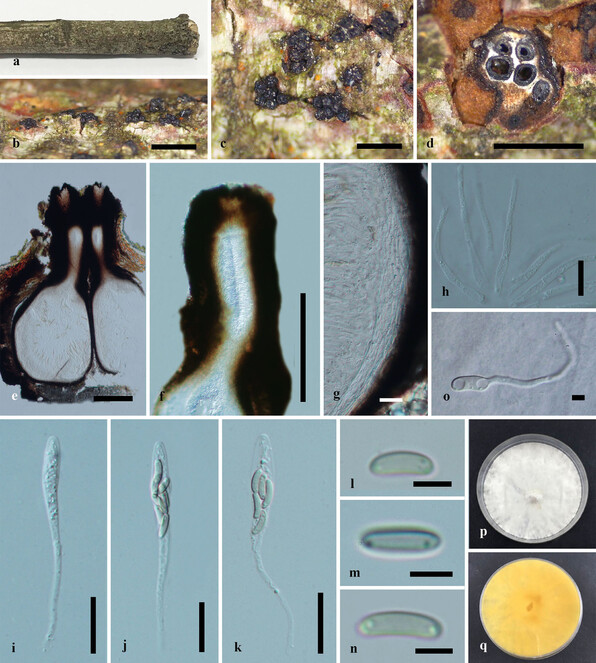
Saprobic on dead twigs of Acer palmatum (Fig.
Paraeutypella citricola (HMAS 290660) a–c. stromata on substrate; d. cross section of stroma; e. vertical section through stroma showing ostiolar necks and perithecia; f. ostiolar canal; g. peridium; h. paraphyses; i–k. asci; l–n. ascospores; o. germinating ascospore; p, q. culture on PDA after 6 weeks from above and below. Scale bars: 500 µm (b–d), 200 µm (e–g), 20 µm (g–l), 5 µm (m–o).
Culture characteristics – Colonies on PDA, reaching 21 mm diam. after 2 weeks at 20–25oC, medium dense, circular to slightly irregular, slightly raised, cottony surface; colony from above: at first white, becoming buff; from below: yellowish-white at margin, yellow to brown at centre; mycelium yellowish.
Notes
Eutypella citricola was described by
Paraeutypella guizhouensis, the type of Paraeutypella, morphologically resembles P. citricola both having immersed stromata, perithecia each with a long ostiolar neck and allantoid, aseptate ascospores with an oil droplet at each end. However, Paraeutypella citricola differs from P. guizhouensis by the number of perithecia within a stroma (4–6 vs. 6–12). A comparison of the ITS 1.0% (6/576) and β-tubulin 1.2% (5/406) between KUMCC 20-0024 and IRAN 2340C, ITS 1.0% (6/576) and β-tubulin 1.0% (5/406) between KUMCC 20-0024 and HVGRF01, HVVIT07 has been made.
Paraeutypella vitis () , comb. nov. ≡ Sphaeria vitis in Schr. Naturf. Ges. Leipzing 1: 39 (1822)
Nomenclature
= Valsa vitis (Schwein.) Fuckel, Jb. Nassau. Ver. Naturk. 23-24: 199 (1870)
= Engizostoma vitis (Schwein.) Kuntze, Revis. Gen. pl. (Leipzig) 3 (3): 475 (1898)
= Eutypella vitis (Schwein.) Ellis & Everh., The North American Pyrenomycetes: 490 (1892)
Notes
Eutypella vitis was collected from young shoots of grape vines in New York and was introduced by
Identification keys
|
Key to species similar to Diatrypella longiasca |
||
| 1 | Ascospores 4–5 μm long on average | Diatrypella major |
| – | Ascospores 6–10 μm long on average | 2 |
| 2 | Entostroma yellowish or olive-green | 3 |
| – | Entostroma white | 4 |
| 3 | Asci larger, 120–150 × 15.5–21.5 μm | D. tectonae |
| – | Asci smaller, 40 × 8–12 μm | D. frostii |
| 4 | Stromata small, up to 2 mm diam. | 5 |
| – | Stromata larger than 2 mm | 6 |
| 5 | 1–4 ascomata per stromata, on twigs of Hevea brasiliensis | D. heveae |
| – | 3–4 ascomata per stromata, on seed pods of Delonix regia | D. delonicis |
| 6 | 4–8 ascomata per stromata, (0.25–0.45 mm) with obscure ostiolar necks | D. vulgaris |
| – | 2–5 ascomata per stromata, (0.5–0.7 mm) with prominent ostiolar necks | D. longiasca |
|
Key to species of Paraeutypella |
||
| 1 | Stromata immersed | Paraeutypella citricola |
| – | stromata erumpent | 2 |
| 2 | Short ostiolar neck and longer asci (55–80 × 5–9 μm) | P. vitis |
| – | Long ostiolar neck and shorter asci (40–46 × 6–8 μm) | P. guizhouensis |
Analysis
Phylogenetic analyses
The combined ITS and β-tubulin matrix comprises 79 sequences that represents the genera in Diatrypaceae including the outgroup taxa. The best scoring RAxML tree is shown (Fig.
Species of Eutypella are polyphyletic in our phylogram, while new isolates KUMCC 20-0023 and KUMCC 20-0024 grouped in a clade that comprises Eutypella citricola Syd. & P. Syd. and Eutypella vitis (Schwein.) Ellis & Everh. (Fig.
Our new strains KUMCC 20-0021 and KUMCC 20-0022 are accommodated within Diatrypella with high statistical support (96% ML, 1.00 BYPP) as a distinct lineage.
Discussion
This study introduces a new genus, Paraeutypella and accepts 22 genera in Diatypaceae. According to the previous analyses of combined ITS and β-tubulin sequence data, the genus Eutypella has been often identified as polyphyletic in Diatrypaceae (
Eutypella citricola groups separately from Eutypella sensu stricto with Eutypella vitis and our newly-generated strains. These new strains are introduced as a new genus, Paraeutypella with three species viz. P. citricola, P. guizhouensis and P. vitis. We studied the morphological characteristics of the species belonging to this clade and found considerable morphological differences from Eutypella sensu stricto. The differences include stromata with 4–25 groups of perithecia in a valsoid configuration, 3–6 sulcate, long ostiolar necks; thus, we consider them to belong in a distinct genus from the Eutypella and hence, we introduce the novel Paraeutypella.
There does not appear to be any host-specificity since members of Diatypaceae are found on a wide range of hosts in various habitats. Diatypaceae species frequently have been identified as saprobes on the decaying wood of angiosperms. Few endophytes, such as Diatrypella frostii Peck and Peroneutypa scoparia (Schwein.) Carmarán & A.I. Romero, have been reported (
In our phylogenetic analyses, some species of Diatrypella: D. favacea (Fr.) Ces. & De Not., D. iranensis Mehrabi & Hemmati, D. macrospora Mehrabi et al. and D. pulvinata Nitschke formed a distinct lineage (Fig.
Acknowledgements
This work was funded by grants of the National Natural Science Foundation of China (NSFC Grants Nos. 31670027 & 31460011). Dr. Shaun Pennycook is thanked for the nomenclatural advice. Nalin N. Wijayawardene gratefully acknowledges Natural Science Foundation of China (grant No. NSFC 31950410558) and grant FAMP201906K provided by the State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University. Dong-Qin Dai thanks the National Natural Science Foundation of China (NSFC 31760013) and the Thousand Talents Plan, Youth Project of Yunnan Provinces for support. Monika C. Dayarathne would like to thank National Natural Science Foundation of China (No. 31972222, 31560489). Lakmali S. Dissanayake would like to thank Ms. D.S. Marasinghe and Ms. S. N. Wijesinghe for valuable suggestions and guidance.
References
- Molecular phylogenetic studies on the Diatrypaceae based on rdna-ITS sequences.Mycologia96(2). https://doi.org/10.2307/3762061
- Heart-rot and associated fungi in Alnus glutinosastands in Latvia.Scandinavian Journal of Forest Research27(4):327‑336. https://doi.org/10.1080/02827581.2012.670727
- Detection of Eutypa lata and Eutypella vitis in grapevine by nested multiplex polymerase chain reaction.Phytopathology97:737‑747. https://doi.org/10.1094/PHYTO-97-6-0737
- Schema di classificazione degli sferiacei italici aschigeri: più omeno appartenenti al genere Sphaerianell’antico significato attribuitogli da Persoo.Commentario della Società Crittogamologica Italiana1:205.
- A new species and a new record of Diatrypaceae from Panama.Mycologia105:681‑688. https://doi.org/10.3852/12-131
- Diatrypasimilis australiensis, a novel xylarialean fungus from mangrove.Mycologia102:430‑437. https://doi.org/10.3852/09-142
- The sooty moulds.Fungal Diversity66:1‑36. https://doi.org/10.1007/s13225-014-0278-5
- Bambusicolous fungi.Fungal Diversity82:1‑105. https://doi.org/10.1007/s13225-016-0367-8
- Halodiatrype, a novel diatrypaceous genus from mangroves with H. salinicola and H. avicenniae spp. nov.Mycosphere7(5):612‑627. https://doi.org/10.5943/mycosphere/7/5/7
- Morpho-molecular characterization of microfungi associated with marine based habitats.Mycosphere11(1):1‑188. https://doi.org/10.5943/mycosphere/11/1/1
- Modern taxonomic approaches toidentifying diatrypaceous fungi from marine habitats, with a novel genusHalocryptovalsa Dayarathne & K.D.Hyde, gen. nov.Cryptogamie, Mycologie41(3). https://doi.org/10.5252/cryptogamie-mycologie2020v41a3
- Taxonomy and molecular phylogeny of Diatrypaceae (Ascomycota, Xylariales) species from the Brazilian semi-arid region, including four new species.Mycological Progress15(6). https://doi.org/10.1007/s11557-016-1194-8
- Diversity and significance of fungal endophytes from living stems of naturalized trees from Argentina.Fungal Diversity41(1):29‑40. https://doi.org/10.1007/s13225-009-0012-x
- Mycologic botany, Phrenomygetes.Published by Ellis and Everhart New Field,New Jersey,490pp.
- Fungal databases, U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/. Accessed on: 2020-12-10.
- Taxonomic notes on Eutypella vitis, Cryptosphaeria populina, and Diatrype stigma.Mycologia79:135‑139. https://doi.org/10.1080/00275514.1987.12025379
- Diatrypaceae in the Pacific Northwest.Mycotaxon20:401‑460.
- Studies in Diatrypaceae: the new species Eutypa microasca and investigation of ligninolytic enzyme production.Sydowia66:99‑114. https://doi.org/10.12905/0380.sydowia66(1)2014-0099
- BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucleic Acids Symposium Series.41:95‑98.
- MRBAYES: Bayesian inference of phylogenetic trees.Bioinformatics17:754‑755. https://doi.org/10.1093/bioinformatics/btg180
- Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa.Fungal Diversity96:1‑242. https://doi.org/10.1007/s13225-019-00429-2
- Refined families of Sordariomycetes.Mycosphere11:305‑1059. https://doi.org/10.5943/mycosphere/11/1/7
- The faces of fungi database: fungal names linked with morphology, molecular and human attributes.Fungal Diversity74:3‑18. https://doi.org/10.1007/s13225-015-0351-8
- MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization.Briefings in Bioinformaticshttps://doi.org/10.1093/bib/bbx108
- Dictionary of the Fungi.10th Edn. CAB International, Wallingford.
- An additional marine fungal lineage in the Diatrypaceae, Xylariales: Pedumispora rhizophorae.Botanica Marina57(5). https://doi.org/10.1515/bot-2014-0017
- A new genus Allodiatrype, five new species and a new host record of diatrypaceous fungi from palms (Arecaceae).Mycosphere11(1):239‑268. https://doi.org/10.5943/mycosphere/11/1/4
- Molecular identification and detection of Eutypa lata in grapevine.Mycological Research109:799‑808. https://doi.org/10.1017/S0953756205002893
- Cryptovalsa ampelina on grapevines in NE Spain: identification and pathogenicity.Phytopathologia Mediterranea45:101‑109. https://doi.org/10.1520/STP37480S
- Species of Diatrypaceae associated with grapevine trunk diseases in Eastern Spain.Phytopathologia Mediterranea51:528‑540. https://doi.org/10.14601/Phytopathol_Mediterr-9953
- Identification and pathogenicity of Botryosphaeriaceae species associated with coast live oak (Quercus agrifolia) decline in southern California.Mycologia105:125‑140. https://doi.org/10.3852/12-047
- Canker disease of willow and poplar caused by Cryptosphaeria pullmanensisrecorded in China.Forest Pathology46(4):327‑335. https://doi.org/10.1111/efp.12261
- A new species and a new record of Diatrypaceae from Iran.Mycosphere6(1):60‑68. https://doi.org/10.5943/mycosphere/6/1/7
- Diatrypella macrospora sp. nov. and new records of diatrypaceous fungi from Iran.Phytotaxa252(1). https://doi.org/10.11646/phytotaxa.252.1.4
- Two new species of Eutypella and a new combination in the genus Peroneutypa (Diatrypaceae).Mycological Progress18:1057‑1069. https://doi.org/10.1007/s11557-019-01503-4
- Creating the CIPRES Science Gateway for inference of large phylogenetic trees.New Orleans, Louisiana, November, 1–8.In Gateway Computing Environments Workshop 2010 (GCE). https://doi.org/10.1109/GCE.2010.5676129
- Pyrenomycetes Germanici Die Kernpilze Deutschlands Bearbeitet von. [Dr. Th. Nitschke].Erster Band,Erste Lieferung i-ii.Germany,Bresalau,156pp. [InVerlag von Eduard Trewendt].
- MrModeltest v.2.2. Program distributed by the author: 2. Evolutionary Biology Centre.Uppsala University 1–2.
- Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium Are Nonorthologous.Molecular Phylogenetics and Evolution7(1):103‑116. https://doi.org/10.1006/mpev.1996.0376
- Occurrence of Eutypella microtheca in grapevine cankers in Mexico.Phytopathologia Mediterranea54:86‑93.
- Liberomyces gen. nov. with two new species of endophytic coelomycetes from broadleaf trees.Mycologia104:198‑210. https://doi.org/10.3852/11-081
- Molecular and morphological evidence for the delimitation of Xylaria hypoxylon.Mycologia101:256‑268. https://doi.org/10.3852/08-108
- Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries.Fungal Diversity 102102:1‑203. https://doi.org/10.1007/s13225-020-00448-4
- Fig.Tree. Tree Fig. Drawing Tool.v. 1.4.0. URL: http://tree.bio.ed.ac.uk/software/fgtree/.
- Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference.Journal of Molecular Evolution43:304‑311. https://doi.org/10.1007/BF02338839
- Taxonomie et nomenclatijre des Diatrypaceae à asques octospores.Mycologia Helvetica2:285‑648.
- A reassessment of the species concept in Eutypa lata, the causal agent of Eutypa dieback of grapevine.Phytopathology96:369‑377. https://doi.org/10.1094/PHYTO-96-0369
- First report of Monosporascus cannonballus on watermelon in Brazil.Plant Disease94(2):278‑278. https://doi.org/10.1094/pdis-94-2-0278b
- Towards unraveling relationships in Xylariomycetidae (Sordariomycetes).Fungal Diversity73:73‑144. https://doi.org/10.1007/s13225-015-0340-y
- Novel taxa of Diatrypaceae from para rubber (Hevea brasiliensis) in northern Thailand; introducing a novel genus Allocryptovalsa.Mycosphere8(10):1835‑1855. https://doi.org/10.5943/mycosphere/8/10/9
- Morpho-molecular characterization of Peroneutypa (Diatrypaceae, Xylariales) with two novel species from Thailand.Phytotaxa356(1). https://doi.org/10.11646/phytotaxa.356.1.1
- Fungi Argentini novi v. critici.Anales Museo Nacional Buenos Aires6:1‑23.
- RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Bioinformatics30(9):1312‑1313. https://doi.org/10.1093/bioinformatics/btu033
- Microfungi on Tamarix.Fungal Diversity82(1):239‑306. https://doi.org/10.1007/s13225-016-0371-z
- Phylogenetic and morphological appraisal of Diatrype lijiangensis sp. nov. (Diatrypaceae, Xylariales) from China.Asian Journal of Mycology2:198‑208. https://doi.org/10.5943/ajom/2/1/10
- Diversity of diatrypaceous fungi associated with grapevine canker diseases in California.Mycologia102(2):319‑336. https://doi.org/10.3852/08-185
- Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia.Fungal Diversity49(1):203‑223. https://doi.org/10.1007/s13225-011-0094-0
- Identification and characterization of Eutypa leptoplaca, a new pathogen of grapevine in Northern California.Mycological Research108:1195‑1204. https://doi.org/10.1017/S0953756204000863
- The genus Cryptosphaeria in the western United States: taxonomy, multilocus phylogeny and a new species, C. multicontinentalis.Mycologia107(6):1304‑1313. https://doi.org/10.3852/15-115
- Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri.Fungal Diversity52:169‑189. https://doi.org/10.1007/s13225-011-0110-4
- Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California.Plant Disease93(6):584‑592. https://doi.org/10.1094/pdis-93-6-0584
- Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota).Molecular Phylogenetics and Evolution98:210‑232. https://doi.org/10.1016/j.ympev.2016.02.010
- Pyrenomycetes of the Great Smoky Mountains National Park. II. Cryptovalsa Ces. et De Not. and Diatrypella (Ces. et De Not.) Nitschke.Fungal Diversity1‑12.
- Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae).Canadian Journal of Microbiology58:54‑56. https://doi.org/10.1139/w11-105
- Diversity and antimicrobial activities of the fungal endophyte community associated with the traditional Brazilian medicinal plant Solanum cernuum Vell. (Solanaceae).Canadian Journal of Microbiology58:54‑66. https://doi.org/10.1139/w11-105
- Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation.Study in Mycology92:135‑154. https://doi.org/10.1016/j.simyco.2018.05.001
- Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In:(Eds.) PCR protocols: a guide to methods and applications.Academic Press,San Diego, California, USA.,315–322pp. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Outline of fungi and fungus-like taxa.Mycosphere11:1060‑1456. https://doi.org/10.5943/mycosphere/11/1/8
- A simplified method for chromosome DNA preparation from filamentous fungi.Mycosystema20:575‑577.
- Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses.MBC genomics3:4. https://doi.org/10.1186/1471-2164-3-4