|
Biodiversity Data Journal : Taxonomic paper
|
|
Corresponding author: Kai Heller (kaiheller@gmx.de)
Academic editor: Vladimir Blagoderov
Received: 02 Sep 2015 | Accepted: 31 Mar 2016 | Published: 01 Apr 2016
© 2016 Kai Heller, Björn Rulik.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Heller K, Rulik B (2016) Ctenosciara alexanderkoenigi sp. n. (Diptera: Sciaridae), an exotic invader in Germany? Biodiversity Data Journal 4: e6460. doi: 10.3897/BDJ.4.e6460
|

|
A new species of the genus Ctenosciara Tuomikoski, 1960 is here described based upon a single specimen, obtained from collectings in the garden at Museum Alexander Koenig in Bonn. Ctenosciara alexanderkoenigi sp. n. differs from all other congeneric European species by its striking coloration and distinct male genitalia. However, DNA barcoding reveals associations with two specimens from New Zealand. Therefore a recent migration of Ctenosciara species from the Australasian Region, the likely center of origin of the genus, is discussed. A key to the European species of Ctenosciara is provided. Barcoding results reveale that Ctenosciara exigua is not clearly distinguished from Ctenosciara hyalipennis by its COI sequence (both share the same BIN BOLD:AAH3983) and that its species status may be questionable.
New species, faunistics, invasive species, DNA barcoding, identification key
Introduction
Ctenosciara was erected by
Materials and methods
The new species was extracted from the catch of a standard malaise trap equipped with a prototype of the Automatic Malaise Trap Changer (AMTC;
Description and specimen deposition
Habitus photos were captured with the aid of a Canon D60 camera fitted with a MP-E 65mm macro photo lens. More detailed close-up photos of specimens were created using a MCA-510 USB microscope camera by TUCSEN (Xintu Photonics Co., Ltd.). Between 15 and 40 images taken at different focal lengths were merged with the aid of the Public Domain Software CombineZP using “Weighted Average” method. All images were retouched using the freely available software GIMP, version 2.8.0. Species descriptions were prepared using DELTA (DEscription Language for TAxonomy) (
Molecular Analysis
G enomic DNA was extracted at ZFMK from the entire specimen using the BioSprint96 magnetic bead extractor by Qiagen (Hilden, Germany). Polymerase chain reaction (PCR) was carried out in total reaction mixes of 20 μl, including 2 μl of undiluted DNA template, 0,8 μl of each primer (10 pmol/μl), 2 μl of ‘Q-Solution’ and 10 μl of ‘Multiplex PCR Master Mix’, containing hot start Taq DNA polymerase and buffers. The latter components are available in the Multiplex PCR kit from Qiagen (Hilden, Germany). PCR reactions were run individually and not multiplexed.
Thermal cycling was performed on GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA, USA) as follows: hot start Taq activation: 15 min at 95°C; first cycle set (15 repeats): 35-s denaturation at 94°C, 90-s annealing at 55°C (−1 °C/cycle) and 90-s extension at 72°C. Second cycle set (25 repeats): 35-s denaturation at 94°C, 90-s annealing at 40°C and 90-s extension at 72°C; final elongation 10 min at 72°C using the primers LCO1490: 5´-GGTCAACAAATCATAAAGATATTGG- 3´ and C1-N-2191 (aka Nancy): 5´-CCCGGTAAAATTAAAATATAAACTTC- 3´ (
Sequencing of the unpurified PCR products in both directions was conducted at Beijing Genomics Institute (Hongkong, CN). Sequence analysis was done using the Geneious® software version 7.1.7 (http://www.geneious.com). All sequences were deposited in BOLD (https://doi.org/10.5883/DS-CTENSCIA) and GenBank under accession numbers KT601633-KT601635 .
Data analysis
Public BOLD API was queried for distribution pattern of Nearctic Sciaridae in order to test for sampling bias (
Taxon treatments
Ctenosciara alexanderkoenigi , sp. n.
- Barcode of Life HRCTE001-15
- ZooBank urn:lsid:zoobank.org:act:1E97DF91-7C19-4C18-9A1C-E4DFC9CB3E5C
-
scientificName: Ctenosciara alexanderkoenigi; genus:Ctenosciara; specificEpithet:alexanderkoenigi; scientificNameAuthorship:Heller & Rulik, 2016; country:Germany; countryCode:DE; stateProvince:North-Rhine-Westphalia; county:Cologne; municipality:Bonn; locality:Museum Koenig; verbatimElevation:67 m; decimalLatitude:50.721944; decimalLongitude:7.113611; samplingProtocol:Malaise trap; eventDate:07/06/2014; startDayOfYear:155; endDayOfYear:159; year:2014; month:6; day:7; habitat:museum´s garden; individualCount:1; sex:male; lifeStage:adult; preparations:slide; catalogNumber:ZFMK-TIS-2527968; recordedBy:Björn Rulik; otherCatalogNumbers:ZFMK-DIP-00011896; institutionCode:ZFMK
Description
Head. Eye bridge 2–3 rows of facets. Antenna with scape and pedicel brightened. LW-index of 4th antennal flagellar segment 2.65; neck 0.35 × the segment width (
Diagnosis
This beautiful species is conspicuous among the European species of Ctenosciara by its eye-catching and contrasting coloration (
Etymology
The new species is named in honour of the founder of the Koenig museum in Bonn, Alexander Koenig (1885-1940).
Distribution
Besides the holotype, the species also appears to be present in New Zealand as confirmed by matching COI sequences on BOLD. We have not studied that material as of yet which is deposited in the Biodiversity Institute of Ontario, Canada.
Taxon discussion
After having seen the conspicuously looking Ctenosciara specimen from the museum's garden in Bonn for the first time, we were convinced of having discovered a new species native to Europe. The yielded COI sequence showed a 7% distance on BOLD to the nearest neighbour from Australia and convinced us furthermore of having an unknown species. After submitting the sequence to BOLD, it was shown to be identical to two other also newly submitted sequences from New Zealand, sharing the same BIN BOLD:ACP7364. Initially having consulted the key to the New Zealand species (
Ctenosciara exigua
- Barcode of Life HRCTE002-15
- Barcode of Life HRCTE003-15
-
scientificName: Ctenosciara exigua; genus:Ctenosciara; specificEpithet:exigua; scientificNameAuthorship:Salmela & Vilkamaa, 2005; country:Finland; countryCode:FI; stateProvince:Lapland; municipality:Enontekiö; locality:Pikkuvaarat SW; verbatimElevation:493; verbatimLatitude:68°07'49.7'' N; verbatimLongitude:24°02'39.8'' E; samplingProtocol:Malaise trap; eventDate:12/09/2014; endDayOfYear:164; year:2014; month:9; day:12; habitat:Poor sedge fen; individualCount:1; sex:male; lifeStage:adult; preparations:slide; catalogNumber:ZFMK-TIS-2544881; recordedBy:Jukka Salmela; otherCatalogNumbers:ZFMK-TIS-2544881; institutionCode:ZFMK
-
scientificName: Ctenosciara exigua; genus:Ctenosciara; specificEpithet:exigua; scientificNameAuthorship:Salmela & Vilkamaa, 2005; country:Finland; countryCode:FI; stateProvince:Lapland; municipality:Savukoski; locality:Tyyroja; verbatimElevation:251; verbatimLatitude:68°09'00" N; verbatimLongitude:28°33'00" E; samplingProtocol:Malaise trap; eventDate:05/08/2014; endDayOfYear:217; year:2014; month:8; day:5; habitat:alpine brook, stony; individualCount:1; sex:male; lifeStage:adult; preparations:slide; catalogNumber:ZFMK-TIS-2544914; recordedBy:Jukka Salmela; otherCatalogNumbers:ZFMK-TIS-2544914; institutionCode:ZFMK
Description
See
Diagnosis
Ctenosciara exigua was described based on several specimens from mires in Central Finland. It was differentiated from Ctenosciara hyalipennis by the evenly broad gonostyli with lacking megasetae at the dorsal side of the apical tooth, the smaller size and less setose CuA2. However, the most distinctive character, the shorter and roundish tegmen, was not mentioned. In Ct. hyalipennis the tegmen is much longer than wide, nearly triangular. In our material, the number of macrotrichia on CuA2 varies from 0 to 18 and the tibial comb was also found to be undivided in some specimens. There were usually differences to the original description in every specimen studied. As seen in
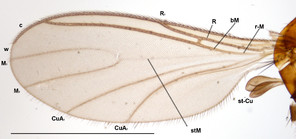
Ctenosciara exigua Salmela and Vilkamaa (2005). Specimen ZFMK-TIS-2544914.
b: Tegmen, scale 0.1 mm
c: Wing, gonostylus, scale 1 mm
d: Tibial comb, scale 0.1 mm
BIN algorithm of BOLD indicates that the COI sequence of Ctenosciara exigua is not significantly different from that of Ctenosciara hyalipennis and belongs to the same BIN BOLD:AAH3983. More than 3000 specimens belonging to that same BIN are recorded from the South West and South East of Canada opposed by only roughly 1000 central-European records. Comparisons based on K2P distances within and between regions show closer affinities of Ctenosciara exigua to the Nearctic population than to the European (
Distribution
Since the original description from Finland, the species was mentioned again by
Taxon discussion
The barcoding results coupled with the fact that Ctenosciara hyalipennis and Ct. exigua (in our understanding) are quite polymorphic raise the question " Is Ctenosciara exigua really a distinct species?" or "Is it only one variant of the former?". In Central Europe, Ct. hyalipennis shows two distinct morphs. The early spring form is larger and has clearly clavate gonostyles, whereas the summer variant is smaller, brighter and the shape of the gonostyles is just as parallel as in Ctenosciara exigua. The summer variant was treated as Ctenosciara thiedei in
Geographic haplotype distribution. ENEA = East Nearctic, FIN = Finland, * = Ct. exigua, GER = Germany, NOR = Norway, WNEA = West Nearctic, see also
| hap |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
| ENEA | 2711 | 213 | 1 | 1 | 1 | |||||||||||||||
| FIN* | 2 | |||||||||||||||||||
| GER | 132 | 2 | 14 | 853 | 1 | 1 | 3 | 3 | 5 | 2 | 4 | 2 | 1 | 2 | 1 | 1 | 1 | |||
| NOR | 1 | 2 | ||||||||||||||||||
| WNEA | 8 | 79 | ||||||||||||||||||
| total | 2854 | 294 | 1 | 1 | 16 | 853 | 1 | 1 | 3 | 3 | 5 | 2 | 4 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
Identification keys
|
Key to the European species of Ctenosciara All currently known European species of Ctenosciara (
|
||
| 1 | Bright, orange-coloured species. Scape and pedicellus yellow. | 2 |
| – | Unicolorous, brownish species. Scape and pedicellus not brightened. | 3 |
| 2 | Body nearly unicolored, orange. CuA2 and stM with macrotrichia. Tibial comb on fore tibia divided. | Ctenosciara lutea (Meigen, 1804) |
| – | Body bicolored, thorax mainly orange, abdomen mainly brown. CuA2 and stM without macrotrichia. Tibial comb on fore tibia undivided. | Ctenosciara alexanderkoenigi sp. n. |
| 3 | Larger species, wing length > 2 mm. CuA2 with more than 10 macrotrichia. Front tibial comb strictly divided. Tegmen conical, longer than broad. | Ctenosciara hyalipennis (Meigen, 1804) |
| – | Smaller species, wing length ≤ 2 mm. CuA2 bare or with less than 8 macrotrichia. Apical comb on fore tibia undivided or unclearly divided. Tegmen roundish, not longer than broad. | Ctenosciara exigua Salmela & Vilkamaa, 2005 |
Discussion
The identification of small Diptera, especially of Sciaridae is based primarally on differences in the male genitalia. In this regard, high quality hand drawings of the hypopygium and the gonostylus at least have become the standard in the description of species. However there are two issues, which led us to depart from this tradition. The first is the sheer number of undescribed species that exist even in a well studied region such as Europe. Thorough drawings tend to be extremely time consuming and thus high quality stacked photos offer a quicker alternative. Secondly, very often, genitalia do not show clear differences at a first glance. DNA barcoding offers a means in which to unravel cryptic diversity and resolve species complexes that might go ignored and/or unnoticed. Interestingly coloration is proving more and more to be an effective tool for taxonomic differentiation in the Sciaridae despite being previously deemed unimportant. Conspecific variation in colouration appears to be minimal in the Sciaridae of the same DNA makeup. When species concepts have to be reconsidered, high quality photo documentation, as is standard in the BOLD project will be a useful method for evaluation. Furthermore a barcode assigned to a BIN will become indispensable to overcome the challenge of future biodiversity issues. We believe, that the rapid description of Ctenosciara alexanderkoenigi coupled with the BDJ reviewing system might be a robust and ground-breaking way to accelerate and stabilize taxonomy in the future.
Acknowledgements
Results presented here were achieved within the framework of the German Barcode of Life, a project of the Humboldt Ring, granted by the German Federal Ministry for Education and Research (German Barcode of Life GBOL1: BMBF #01LI1101A). Additionally, we would like to thank Laura von der Mark und Jana Thormann for their technical assistance in the lab. We also thank Werner Mohrig (Poseritz, Germany) for letting us compare our specimen with the type material of Ctenosciara nigrostyla and to the German Entomological Institute (Müncheberg) for letting us use their photographic equipment. Vladimir Blagoderov (London, United Kingdom), Rob Deady (Cork, Ireland), Arne Köhler (Müncheberg, Germany), Jukka Salmela (Turku, Finland) and Pekka Vilkamaa (Helsinki, Finland) improved by their useful comments an earlier version of the manuscript. Additional thanks to Rob Deady for the language check.
References
-
Phylogeny in cryptic weevils: molecules, morphology and new genera of western Palaearctic Cryptorhynchinae (Coleoptera: Curculionidae).Invertebrate Systematics22(5):503‑522. DOI: 10.1071/is07057
-
User’s guide to the DELTA Editor. URL: http://delta-intkey.com
-
MUSCLE: multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Research32(5):1792‑1797. DOI: 10.1093/nar/gkh340
-
DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates.Molecular Marine Biology and Biotechnology3:294‑299.
-
An annotated check list of Swedish black fungus gnats (Diptera, Sciaridae).Sahlbergia15:23‑51.
-
Park und Garten des Museum Alexander Koenig im Wandel der Zeit.Koenigiana6(2):81‑99.
-
The Mycetophilidae of North America. Part IV (Conclusion). Bulletin of the.Maine Agricultural Experimental Station.200.
-
Two new Neuratelia Rondani (Diptera, Mycetophilidae) species from Western Palaearctic: a case of limited congruence between morphology and DNA sequence data.ZooKeys496:105‑129. DOI: 10.3897/zookeys.496.9315
-
DnaSP v5: a software for comprehensive analysis of DNA polymorphism data.Bioinformatics25(11):1451‑1452. DOI: 10.1093/bioinformatics/btp187
-
Beiträge zur Insektenfauna der DDR: Diptera - Sciaridae.Beiträge zur Entomologie40(2):301‑400.
-
Die Trauermücken (Diptera: Sciaridae) von Papua-Neuguinea. Teil III – Gattungen Ctenosciara und Pseudolycoriella.Studia dipterologica20:123‑168.
-
Sciarid flies (Diptera, Sciaridae) of New Zealand. Studia dipterologica.Supplement7:1‑101.
-
Revision of the Black Fungus Gnats (Diptera: Sciaridae) of North America.Studia dipterologica19:141‑286.
-
A revision of the genus Sciara of the family Mycetophilidae (Diptera). Annals of the Entomological Society of.America11:319‑343. DOI: 10.1093/aesa/11.4.319
-
AMTC: Automated Malaise Trap Changer. In: Dorchin N, Kotrba M, Mengual X, Menzel F (Eds)8th International Congress of Dipterology – Abstract volume.Potsdam,440pp. [ISBN978-3-932795-36-7].
-
Sciaridae (Diptera) from central Finland: faunistics and taxonomy.Entomologica Fennica16:287‑300.
-
Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers.Annals of Entomological Society of America87:651‑701. DOI: 10.1093/aesa/87.6.651
-
A taxonomic study on the genus Ctenosciara (Insecta: Diptera: Sciaridae) from Japan.Species Diversity8:119‑131.
-
MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0.Molecular Biology and Evolution30(12):2725‑2729. DOI: 10.1093/molbev/mst197
-
Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve.Biodiversity Data Journal3:e6313. DOI: 10.3897/bdj.3.e6313
-
Untersuchungen über die Arthropodenfauna in Fichtenforsten (Populationsökologie, Energieumsatz). Zoologische Jahrbücher, Abteilung für Systematik, Ökologie und Geographie der.Tiere104:137‑202.
-
Zur Kenntnis der Sciariden (Dipt.) Finnlands. Annales Zoologici Societatis Zoologicae Botanicae Fennicae.“Vanamo”21:1‑164.
-
The genus Ctenosciara Tuomikoski (Diptera, Sciaridae) in New Caledonia, with the description of eight new species.Zootaxa3255:37‑51.
-
The genus Ctenosciara Tuomikoski in China, with descriptions of three new species (Diptera, Sciaridae).Zootaxa2560:42‑50.
Supplementary materials
Download file (13.97 MB)
Download file (2.60 MB)
Download file (32.47 MB)
Download file (9.28 kb)
Download file (10.58 kb)
Download file (102.27 kb)