|
Biodiversity Data Journal :
Software Description
|
|
Corresponding author:
Academic editor: Pavel Stoev
Received: 05 Apr 2016 | Accepted: 14 May 2016 | Published: 01 Nov 2016
© 2016 Kleoniki Keklikoglou, Sarah Faulwetter, Eva Chatzinikolaou, Nikitas Michalakis, Irene Filiopoulou, Nikos Minadakis, Emmanouela Panteri, George Perantinos, Alexandros Gougousis, Christos Arvanitidis
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Keklikoglou K, Faulwetter S, Chatzinikolaou E, Michalakis N, Filiopoulou I, Minadakis N, Panteri E, Perantinos G, Gougousis A, Arvanitidis C (2016) Micro-CTvlab: A web based virtual gallery of biological specimens using X-ray microtomography (micro-CT). Biodiversity Data Journal 4: e8740. https://doi.org/10.3897/BDJ.4.e8740
|
|
Abstract
Background
During recent years, X-ray microtomography (micro-CT) has seen an increasing use in biological research areas, such as functional morphology, taxonomy, evolutionary biology and developmental research. Micro-CT is a technology which uses X-rays to create sub-micron resolution images of external and internal features of specimens. These images can then be rendered in a three-dimensional space and used for qualitative and quantitative 3D analyses. However, the online exploration and dissemination of micro-CT datasets are rarely made available to the public due to their large size and a lack of dedicated online platforms for the interactive manipulation of 3D data. Here, the development of a virtual micro-CT laboratory (Micro-CTvlab) is described, which can be used by everyone who is interested in digitisation methods and biological collections and aims at making the micro-CT data exploration of natural history specimens freely available over the internet.
New information
The Micro-CTvlab offers to the user virtual image galleries of various taxa which can be displayed and downloaded through a web application. With a few clicks, accurate, detailed and three-dimensional models of species can be studied and virtually dissected without destroying the actual specimen. The data and functions of the Micro-CTvlab can be accessed either on a normal computer or through a dedicated version for mobile devices.
Keywords
micro-CT, virtual lab, 3D visualisation, virtual galleries, cyberspecimens, digitised collections
Introduction
X-ray microtomography (micro-CT) is a non-destructive X-ray imaging technology which creates high-resolution three-dimensional data. In the past years, micro-CT has been increasingly used in several biological research fields, such as taxonomy (e.g.
In an increasingly computerised world, the digitisation and dissemination of biodiversity information is growing rapidly, and specialised repositories for a variety of data types exist (e.g. NCBI databases for molecular data, OBIS and GBIF for biogeographical data, Biodiversity Heritage Library for digitised literature, Morpho-D-Base for media files, or MorphoSource for 3D data). However, although many micro-CT datasets of biological specimens are being created and published (e.g.
The Micro-CT virtual laboratory (Micro-CTvlab) was created to contribute to the abovementioned needs. The Micro-CTvlab offers the user a collection of virtual 3D specimens which are annotated with metadata and can be interactively displayed and retrieved through a web-based application. It allows the user to search for adequate datasets and to interact with the 3D models by using a series of online tools. These developments were supported by two complementary research projects: a) the EU FP7 project SYNTHESYS3, in the framework of which protocols, workflows and data management practices as well as online dissemination methods for virtual specimens were researched and developed, and b) the ESFRI LifeWatchGreece infrastructure, which supported the integration of the data into a large semantic infrastructure and provided the technical infrastructure and developments for the Micro-CTvlab.
Project description
Data creation, processing and publication within the Micro-CT vlab
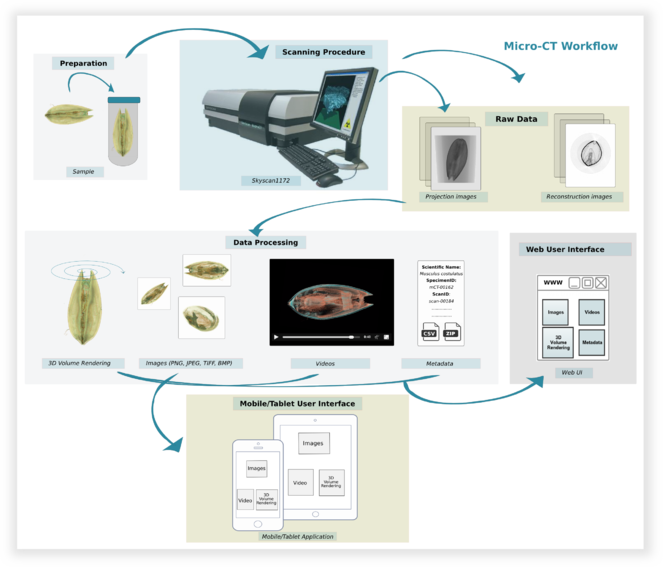
The implementation of the Micro-CTvlab takes into account all steps and procedures which contribute to the creation of the final dataset, from the preparation of the specimen to scanning parameters and post-processing of the data (Fig.
Micro-CTvlab Feature Description
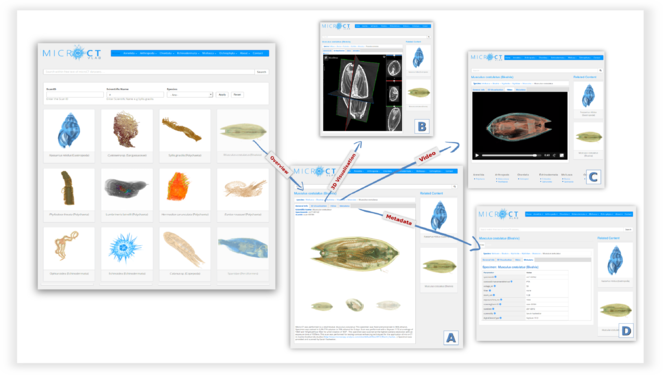
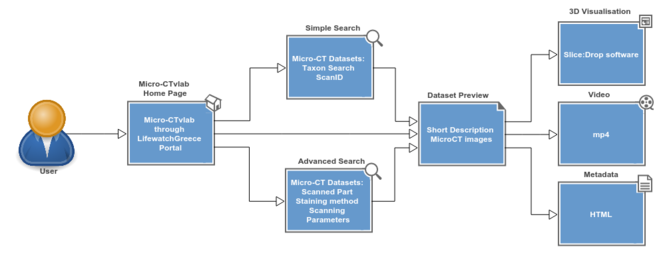
The Micro-CTvlab is a web application compatible with all major web browsers, and available also as a mobile version. So far, 17 micro-CT datasets have been published, representing a selection of marine species scanned with different parameters. On the main page, scans are presented as a preview of images accompanied by the title of the dataset (Fig.
The General Info tab contains a description of the dataset, the taxonomic classification of the specimen and a series of 3D images which serve as a preview to the dataset (A).
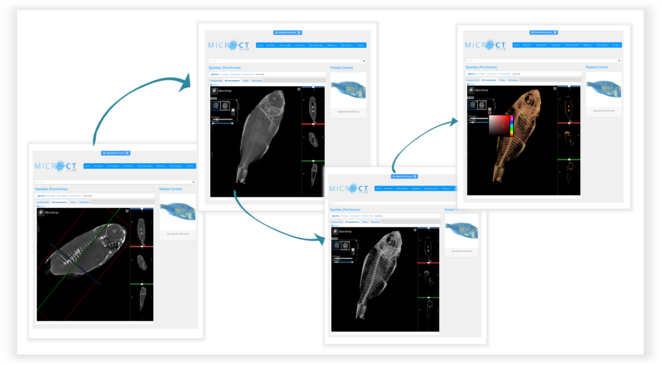
Using the 3D visualisation tab (Fig.
The Video tab displays a short preview video as a demonstration of the specific micro-CT dataset (Fig.
The Metadata tab contains additional information about the dataset, such as contrast enhancement methods, scanning parameters or the creator of the dataset (Fig.
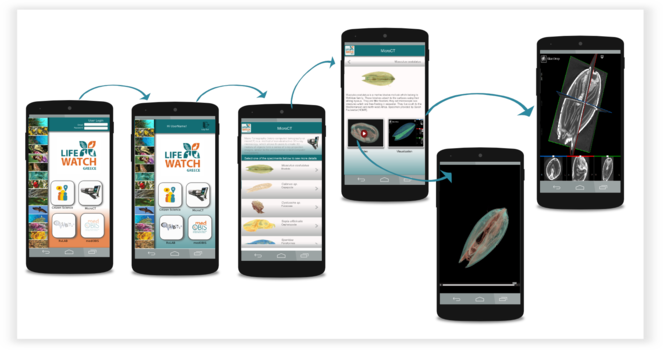
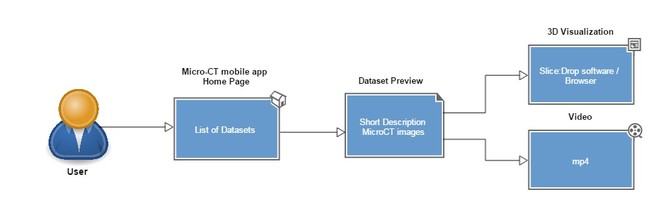
The Micro-CT mobile/tablet application has functionalities similar to the Micro-CTvlab web application but presents these in a simplified and compact version (Fig.
Minimum system requirements
The Micro-CTvlab web application has been tested and works on all modern browsers: Google Chrome version 48 > / Mozilla Firefox version 44 / Safari version 9 / Opera version 34 / Microsoft Edge 25 / Internet Explorer 11.
To use the interactive 3D software Slice:Drop, the following are required:
- Operating System: Windows XP SP3 and newer / Mac OSX 10.6 and newer / Linux / Chrome OS
- Computer speed and processor: Use a computer with a minimum of 1GB of RAM and a 2GHz processor
- Internet Speed: Along with compatibility and web standards, Canvas has been carefully crafted to accommodate low bandwidth environments. A minimum of 512kbps is recommended.
The mobile application requires iOS 7 and newer or Android 2.3 and newer.
This research has been supported by the LifeWatchGreece infrastucture (ESFRI, MIS 384676), funded by the Greek Government under the General Secretariat of Research and Technology (GSRT), National Strategic Reference Framework (NSRF) and the EU FP7 programme SYNTHESYS3 (FP7-312253).
Web location (URIs)
Technical specification
Usage rights
The source code for the mobile application is licensed under MIT license. For the web application the source code is licensed under the GNU General Public License. The content of the MicroCTvlab is available under a Creative Commons Attribution License (CC-BY) unless indicated otherwise (see LifeWatchGreece Data Sharing Agreement).
Implementation
Implements specification
In order to deploy a web application for scientists that could handle the complex requirements (implementing 3D volume rendering software, Web Services and online media presentation) of a development like the Micro-CTvlab, a custom framework was required that can provide a wide range of features for creating and editing content in a simple and user friendly way. Furthermore, open source technologies that could implement these needs were given priority over commercial solutions.
Technical architecture
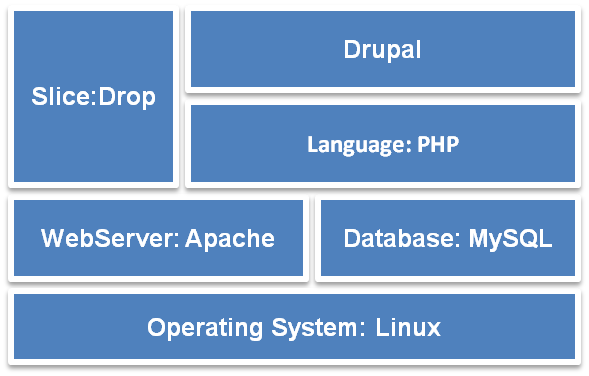
The Micro-CTvlab web application has been developed using open source technologies. It is based primarily on the content management system (CMS) Drupal (v.7) and its software stack is illustrated in Fig.
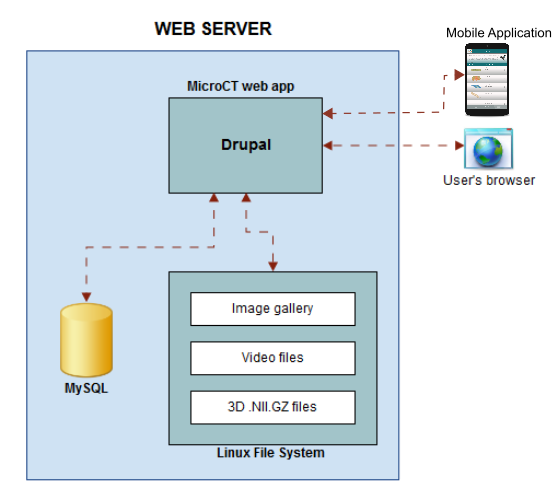
When importing a new dataset, the Micro-CTvlab stores three types of media files (video, images and .nifti files) in the file system and some relevant information in the database (Fig.
For the mobile application development, the Unity3D Platform solution has been adopted. The data presented in the mobile application are acquired by https requests to the Drupal portal (Fig.
Audience
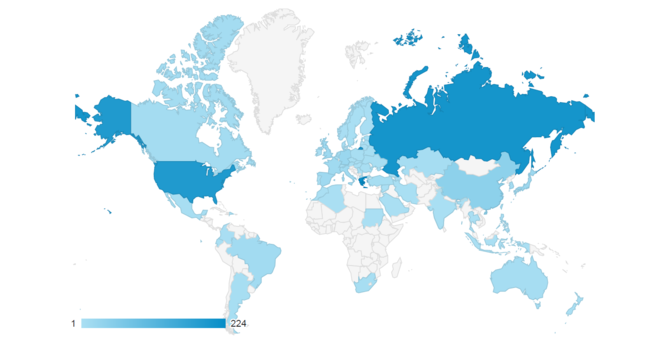
The Micro-CTvlab targets a wide audience which includes academics, scientists, students, artists and animators, as well as everyone who is interested in digitisation methods and biological collections or simply wants to explore the 3D visualisations. Already, the tool has attracted a lot of attention after the first months after its launch (Fig.
Experts in micro-CT technology can use the information in the Micro-CTvlab to compare or discover protocols and scanning parameters (best staining solution, usage of filter, scanning voltage, etc.) for different species. Furthermore, members of the scientific community who are not yet familiar with this technology but work in areas such as taxonomy, evolutionary, developmental or functional biology could be attracted by the Micro-CTvlab since this virtual service presents, through a range of examples, the potential for micro-CT imaging in many research fields.
Natural history museums will naturally be highly interested in the Micro-CTvlab and the underlying technology, since there is a need for massive digitisation and dissemination of natural history collections and this virtual lab could be used as a tool to achieve this. Furthermore, the Micro-CTvlab can be used for educational purposes since it offers information on the morphology and anatomy of species and the 3D model scan be interactively manipulated by the students. The simplified mobile/tablet application could attract an even wider audience who are not experts in any of the abovementioned fields but are interested in exploring biological specimens. Several micro-CT scans can be freely downloaded via the Dryad repository and can be used to create 3D surface models which can then be printed in a 3D printer and thus offer an even more tangible experience regarding the anatomy of specimens. All the remaining micro-CT datasets will be available for downloading in the near future (see paragraph "Conclusions and Future Scenario"). Target user groups include museums, aquariums, herbariums, universities and schools, environmental education associations, research institutes, as well as any member of the general public with an interest in natural sciences.
Additional information
Conclusions and Future Scenario
The Micro-CTvlab is a web-based virtual lab and a mobile application which presents virtual galleries of 3D micro-CT datasets of biological specimens and innovative online tools for their manipulation and exploration. This is, to our knowledge, an important step towards the massive creation, manipulation and dissemination of three-dimensional morphological datasets. We have developed a standardised workflow for the creation of micro-CT datasets, protocols and terms for documenting each dataset with metadata, and a web-based environment for the publication, dissemination and on-the-fly rendering manipulation of these datasets and their metadata. The Micro-CTvlab in its current stage provides a fully-fledged virtual research enviroment integrating all the above steps. However, developments are ongoing to improve several aspects of the current implementation: a) the large size of the micro-CT datasets constitutes a restriction for native integration and upload of the raw data (i.e. the high-resolution cross-section datasets). Currently, several datasets are available only through external links to the Dryad data repository, but an installation of a storage area network (SAN) is planned to overcome this restriction. With this SAN in place, all datasets will be made available for download; b) a service needs to be developed to allow other micro-CT users to submit and share their raw data through the Micro-CTvlab; c) the communication with the LifeWatchGreece data services catalogue needs to be improved to allow refined querying for datasets; d) the process of creating preview files, descriptions and .nifti files for online manipulation needs to be automated so that the integration of additional datasets can be achieved more quickly.
In its current version the Micro-CTvlab is only an initial step for the implementation of the massive digitisation of micro-CT datasets, but it forms the basis for future developments of centralised repositories for such data. The digitisation of natural history collections is rapidly advancing (
In the long term these develoments will transform taxonomic research into a true cyber-discipline: more and more morphological and anatomical data will become available in an electronic, shareable format. This will speed up systematic research, as morphological, high-resolution information can be accessed at the click of a mouse, and analysed in completely new ways by pattern recognition algorithms, steering comparative morphology into a new direction and shifting from the current use of phenetics to that of phenomics (
Acknowledgements
The Micro-CTvlab has been supported by the LifeWatchGreece infrastucture (ESFRI, MIS 384676), funded by the Greek Government under the General Secretariat of Research and Technology (GSRT), National Strategic Reference Framework (NSRF) and the EU FP7 programme SYNTHESYS3 (FP7-312253). The Skyscan 1172 microtomograph has been purchased and installed in the HCMR premises with support from the project MARBIGEN (FP7-264089). The reviewers (Dr Brian Metscher, Dr Nesrine Akkari and one anonymous reviewer) are thanked for their suggestions to improve the manuscript.
Author contributions
Kleoniki Keklikoglou, Sarah Faulwetter, Eva Chatzinikolaou and Christos Arvanitidis designed and planned the concept of the Micro-CTvlab. Kleoniki Keklikoglou, Sarah Faulwetter and Eva Chatzinikolaou created the scans, videos and images. Kleoniki Keklikoglou and Sarah Faulwetter developed and tested workflows, protocols and metadata terms. Nikitas Michalakis and George Perantinos developed the web-based version of the Micro-CTvlab. Irene Filiopoulou and Emmanouela Panteri developed the mobile version of the Micro-CTvlab. Nikos Minadakis was responsible for the LifeWatchGreece metadata catalogue. Alexandros Gougousis developed the LifeWatchGreece portal. All authors contributed to the writing of the manuscript.
References
- A New Dimension in Documenting New Species: High-Detail Imaging for Myriapod Taxonomy and First 3D Cybertype of a New Millipede Species (Diplopoda, Julida, Julidae).PLOS ONE10(8):e0135243. https://doi.org/10.1371/journal.pone.0135243
- Bringing collections out of the dark.ZooKeys209:1‑6. https://doi.org/10.3897/zookeys.209.3699
- Predicting muscle activation patterns from motion and anatomy: modelling the skull of Sphenodon (Diapsida: Rhynchocephalia).Journal of The Royal Society Interface7(42):153‑160. https://doi.org/10.1098/rsif.2009.0139
- First steps towards the development of an integrated metadata management system for biodiversity-related micro-CT datasets.Bruker microCT 2015 User Meeting,Bruges, Belgium.11pp.
- Micro-computed tomography: Introducing new dimensions to taxonomy.ZooKeys263:1‑45. https://doi.org/10.3897/zookeys.263.4261
- Sine Systemate Chaos? A Versatile Tool for Earthworm Taxonomy: Non-Destructive Imaging of Freshly Fixed and Museum Specimens Using Micro-Computed Tomography.PLoS ONE9(5):e96617. https://doi.org/10.1371/journal.pone.0096617
- Slice: drop: Collaborative medical imaging in the browser.ACM SIGGRAPH 2013 Computer Animation Festival,1pp. https://doi.org/10.1145/2503541.2503645
- Phenomics: the next challenge.Nature Reviews Genetics11(12):855‑866. https://doi.org/10.1038/nrg2897
- A dataset describing brooding in three species of South African brittle stars, comprising seven high-resolution, micro X-ray computed tomography scans.GigaScience4(1):52. https://doi.org/10.1186/s13742-015-0093-2
- A dataset comprising four micro-computed tomography scans of freshly fixed and museum earthworm specimens.GigaScience3(1):6. https://doi.org/10.1186/2047-217x-3-6
- Drishti: a volume exploration and presentation tool.8506.Developments in X-Ray Tomography VIII.Spie Proceedings,10pp. https://doi.org/10.1117/12.935640
- Embryonic shell formation in the snail Biomphalaria glabrata: a comparison between scanning electron microscopy (SEM) and synchrotron radiation micro computer tomography (SR CT).Journal of Molluscan Studies74(1):19‑26. https://doi.org/10.1093/mollus/eym044
- MicroCT for developmental biology: A versatile tool for high‐contrast 3D imaging at histological resolutions.Developmental Dynamics238(3):632‑640. https://doi.org/10.1002/dvdy.21857
- Functional morphology of Tethya species (Porifera): 1. Quantitative 3D-analysis of Tethya wilhelma by synchrotron radiation based X-ray microtomography.Zoomorphology125(4):209‑223. https://doi.org/10.1007/s00435-006-0021-1
- The evolution of mammal-like crocodyliforms in the Cretaceous Period of Gondwana.Nature466(7307):748‑751. https://doi.org/10.1038/nature09061
- The pros and cons of using micro-computed tomography in gross and microanatomical assessments of polychaetous annelids.Memoirs of Museum Victoria71:237‑246.
- Sensory epithelia of the fish inner ear in 3D: studied with high-resolution contrast enhanced microCT.Frontiers in Zoology10(1):63. https://doi.org/10.1186/1742-9994-10-63
- Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data.Biodiversity Data Journal1:e1013. https://doi.org/10.3897/bdj.1.e1013
- Description of a soft-bodied invertebrate with microcomputed tomography and revision of the genus Chtonobdella (Hirudinea: Haemadipsidae).Zoologica Scriptan/a:n/a‑n/a. https://doi.org/10.1111/zsc.12165
- Micro-CT scanning provides insight into the functional morphology of millipede genitalia.Journal of Zoology287(2):91‑95. https://doi.org/10.1111/j.1469-7998.2011.00892.x