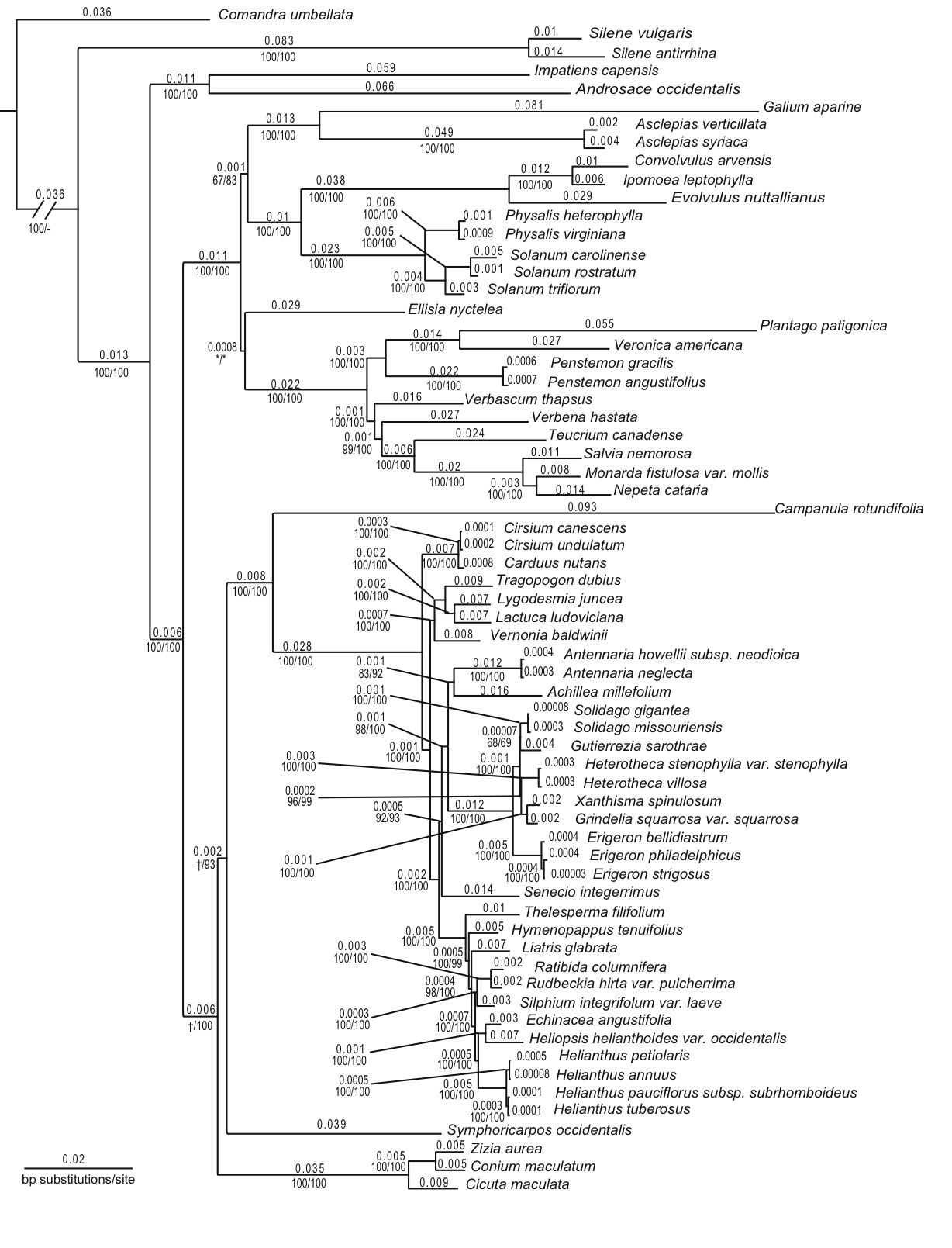
Maximum likelihood (ML) tree (-ln L=46268.63) inferred from the concatenation of 76 plastid, six mitochondrial, and three nuclear ribosomal repeat regions (cpmtnuc; Suppl. material

|
||
|
Maximum likelihood (ML) tree (-ln L=46268.63) inferred from the concatenation of 76 plastid, six mitochondrial, and three nuclear ribosomal repeat regions (cpmtnuc; Suppl. material |
||
| Part of: Aust S, Ahrendsen D, Kellar P (2015) Biodiversity assessment among two Nebraska prairies: a comparison between traditional and phylogenetic diversity indices. Biodiversity Data Journal 3: e5403. https://doi.org/10.3897/BDJ.3.e5403 |