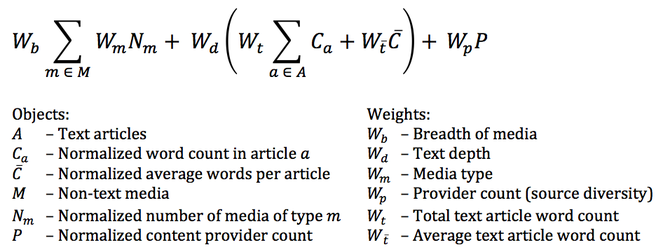|
Biodiversity Data Journal :
Software description
|
The Encyclopedia of Life v2: Providing Global Access to Knowledge About Life on Earth
|
Corresponding author:
Academic editor: Edward Baker
Received: 19 Mar 2014 | Accepted: 24 Apr 2014 | Published: 29 Apr 2014
© 2014 Cynthia Parr, Nathan Wilson, Cynthia Parr, Patrick Leary, Katja Schulz, Kristen Lans, Lisa Walley, Jennifer Hammock, Anthony Goddard, Jeremy Rice, Marie Studer, Jeffrey Holmes, Robert Corrigan, Jr.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Parr C, Wilson N, Leary P, Schulz K, Lans K, Walley L, Hammock J, Goddard A, Rice J, Studer M, Holmes J, Corrigan Jr. R (2014) The Encyclopedia of Life v2: Providing Global Access to Knowledge About Life on Earth. Biodiversity Data Journal 2: e1079. https://doi.org/10.3897/BDJ.2.e1079
|
|
Abstract
The Encyclopedia of Life (EOL, http://eol.org) aims to provide unprecedented global access to a broad range of information about life on Earth. It currently contains 3.5 million distinct pages for taxa and provides content for 1.3 million of those pages. The content is primarily contributed by EOL content partners (providers) that have a more limited geographic, taxonomic or topical scope. EOL aggregates these data and automatically integrates them based on associated scientific names and other classification information. EOL also provides interfaces for curation and direct content addition. All materials in EOL are either in the public domain or licensed under a Creative Commons license. In addition to the web interface, EOL is also accessible through an Application Programming Interface.
In this paper, we review recent developments added for Version 2 of the web site and subsequent releases through Version 2.2, which have made EOL more engaging, personal, accessible and internationalizable. We outline the core features and technical architecture of the system. We summarize milestones achieved so far by EOL to present results of the current system implementation and establish benchmarks upon which to judge future improvements.
We have shown that it is possible to successfully integrate large amounts of descriptive biodiversity data from diverse sources into a robust, standards-based, dynamic, and scalable infrastructure. Increasing global participation and the emergence of EOL-powered applications demonstrate that EOL is becoming a significant resource for anyone interested in biological diversity.
Introduction
Biodiversity science has produced hundreds, if not thousands, of isolated database resources (
The Encyclopedia of Life (EOL, eol.org) is an online database aiming to document all life on Earth. Globally and taxonomically comprehensive, EOL serves descriptive information and media (images, videos, sounds, maps) about biological organisms. While the modern concept of EOL was proposed by E. O. Wilson (
EOL’s focus on description and illustration complements several related global efforts. The Catalogue of Life Partnership (CoL,
The task of documenting all life is vast, perhaps too vast for the relatively small community of formally-trained biodiversity experts (
EOL’s first phase established a basic content aggregation and curation infrastructure with the original website launching in 2008 (
In this paper, we review recent developments added for Version 2 and subsequent releases through Version 2.2. We outline the core features and technical architecture of the system. We summarize milestones achieved so far, both to present results of the system implementation and to establish baselines upon which to judge future improvements and comparisons with other systems. Finally, we discuss the significance of the Encyclopedia of Life to the landscape of biodiversity informatics.
Project description
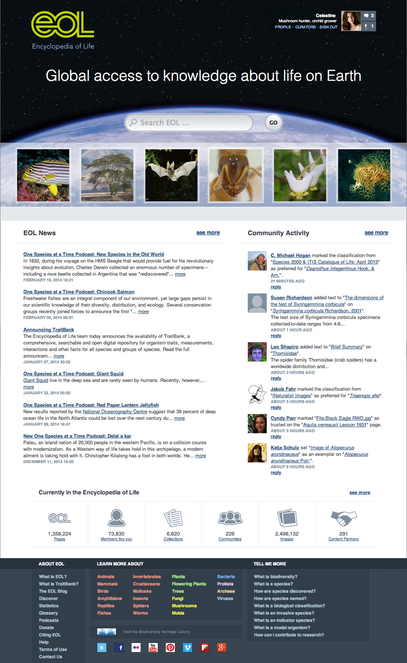
EOL Version 2 involved a complete redesign of page styles to be more personal and engaging. In addition to the “March of Life” (a changing set of images linked to selected EOL pages), the homepage (http://eol.org, Fig.
EOL v2 Homepage. When a member is logged in, the upper right corner of the page features links to member profile, personalized newsfeeds, and other information. Below the site search box, the "March of Life" thumbnails provide links to a sample of taxon pages drawn at random from pages above a minimal richness threshold. Two columns then feature EOL-related news items and an overview of recent community activity, followed by selected site statistics with a link to more detailed statistics over time. The site footer provides quick access to gateway pages (see below) and other site documentation.
Commenting by users was available in the first version of EOL, but it has become a more central feature in EOL Version 2. Comments are now displayed much more prominently and are incorporated into EOL Newsfeeds, which also aggregate user actions relevant to the topic of the newsfeed. Newsfeed topics include users, taxa, collections and communities. EOL members (users who register for accounts on the site) are notified of responses to their comments and actions, and email notifications from newsfeeds can be customized in a preferences panel. The new EOL commenting system resulted in a roughly 4-fold increase in the rate of commenting compared to Version 1.
With the addition of a WYSIWYG editor to the existing text contribution interface, the authoring of taxon descriptions in the EOL interface has become easier in Version 2, and over 7,000 articles have been contributed in this way. In addition, we have introduced a link object so that contributors can submit well-described links to external resources; these are found on the Resources tab.
EOL Version 2 introduced the ability for members to form communities and build collections (of taxa, of image objects, of other collections, etc.) on EOL, as described more fully below (Implementation). EOL collections allow users to collaborate on projects and to annotate and arrange EOL content from a personal point of view. Since the content of collections is available through the EOL API (see Application Programming Interface section below), they can be employed to organize EOL content for use by other applications. This collection-making facility likely is the most powerful new EOL feature; users can add value to the content by organizing it, and software developers can build on this value.
Most of the 1.9 million species described by science (
The EOL Version 2 redesign included a complete rewrite of EOL’s presentation layer with the goal of delivering content in meaningful ways to the widest possible audience regardless of the recipient's device, ability or location. The structure, style and client-side behavior components of each page were separated and rewritten using progressive enhancement techniques (
Accessibility and search engine optimization
Design and architectural changes meet the World Wide Web Consortium (W3C) recommended Web Content Accessibility Guidelines (WCAG) 2.0 (
Internationalization and localization
In partnership with Bibliotheca Alexandrina, the EOL interface system (menus, controls, feedback messages, etc.) was fully internationalized. This work, combined with the separation of structure and style, has allowed the site to support the right to left layout needed for some non-Latin languages such as Arabic. Translation of site elements into specific languages was often accomplished by EOL global partners, e.g. Spanish by Costa Rica's Instituto Nacional de Biodiversidad and Simplified Chinese by the Chinese Academy of Sciences. The abstraction of interface strings has also enabled a partnership with the TranslateWiki platform, which supports interface translation by volunteers into over 120 languages. Once a language reaches a translation threshold of 75%, it is added to the menu of supported languages on EOL. This process has resulted in support of 16 languages in addition to English, with active development continuing for several more. EOL currently displays only text object content that matches a user's preferred language setting, but provides links to content available in other languages. Following these links changes the language setting. The goal is to avoid rendering multiple languages on the same page.
To better support beginning users, EOL now provides pages on general topics such as “What is biodiversity?” and introductory pages to major groups of organisms. Some of these pages are adapted from partner projects such as the Encyclopedia of Earth or the Animal Diversity Web (
Support was provided by John D. and Catherine T. MacArthur Foundation (93466-0 amendment to grant 06-89123-000-GEN), Alfred P. Sloan Foundation (2009-6-076), Smithsonian Institution, Marine Biological Laboratory, and Harvard University.
Technical specification
Repository
Usage rights
Third-party content copyright remains with rightsholders. All content is either in the public domain or licensed for re-use with Creative Commons licenses. All but non-derivative ND licenses are accepted for third-party content (see EOL Policy). User-generated content (e.g. comments, annotations in collections) is CC-BY licensed according to the Community Conditions and Comment policy. All EOL-generated source code is available under the MIT License.
Implementation
Implements specification
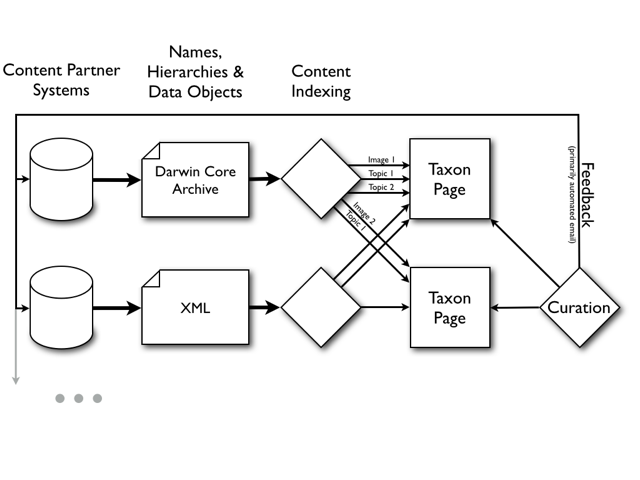
Fig.
High-level data flow from content partners into EOL. Content partners make available EOL data transfer documents (resource documents) that are periodically indexed by EOL. These provide names, name hierarchies, and associated data objects to EOL. EOL aggregates these data and presents them on Taxon Pages. The content assigned to a Taxon Page can be reviewed, hidden, or reassigned to other Taxon Pages by EOL Curators.
Resource documents made available by content partners define the text and multimedia being provided as well as the taxa to which the content refers, the associations between content and taxa, and the associations among taxa (i.e. taxonomies). Expert taxonomists often disagree about the best classification for a given group of organisms, and there is no universal taxonomy for partners to adhere to (
This taxonomic reconciliation process involves comparing the preferred scientific names, synonymy, and taxonomy from an incoming resource document to the same information from all previously indexed resources. It is designed to merge taxa based on synonymy (for example when the preferred name of one taxon is in the synonymy of another) and keep taxa separated that are homonyms (the same scientific name appearing in two distinctly different clades like Morus which is a genus of both birds and plants). Rank information is important to the reconciliation process as it permits the differentiation of cross-rank homonyms. For example, there is a genus of seaweed known as Vertebrata and the same name is used for the group of all organisms with backbones. Reconciliation is an automated process and can make incorrect decisions, so there is a series of operations EOL curators can perform to manually resolve taxonomic and typographic inconsistencies. Ultimately, multiple taxonomic views indexed by EOL are displayed in the Names Tab of a Taxon Page, and EOL curators can choose a preferred taxonomy to display for browsing on the Overview tab.
Partners can provide common names and synonyms as part of their taxon definitions. Synonyms are used by EOL to help determine which taxon definitions should be aggregated into the same Taxon Pages. They are also valuable search keywords that help users find the pages they are looking for.
Previous studies suggest that common names are often more valuable for search than scientific names or synonyms (
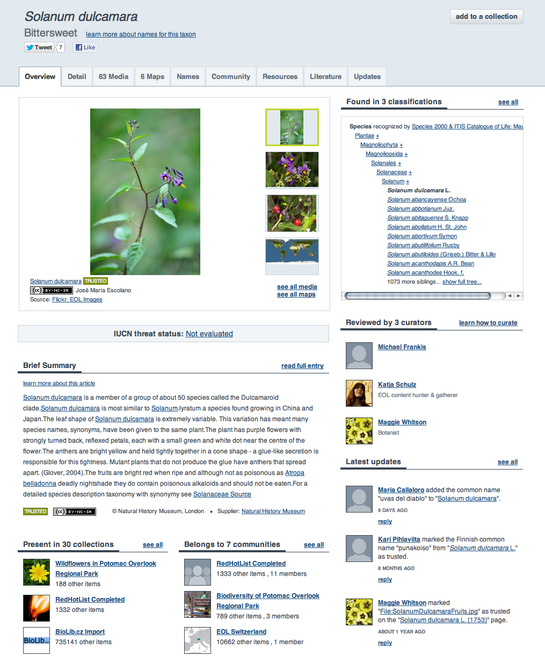
Taxon Pages are the main organizational unit of EOL, presenting a standardized page for every taxonomic entity that the system recognizes. Each Taxon Page has 9 tabs: Overview, Details, Media, Maps, Names, Community, Resources, Literature, and Updates, plus an additional tab for EOL curators, Worklist. The default tab, Overview (Fig.
The Community tab offers information about what EOL Communities and Collections are interested in the taxon, and who the curators of the taxon have been. The Updates tab lists all of the comments on the Taxon Page as well as statistics about the content on the page, including the page’s Richness Score (see Richness score below).
Images, text, videos, sound files, and maps provided by content providers and EOL members are referred to as “Data Objects”. Data Objects are the building blocks of EOL. Taxon Pages are populated through the aggregation of relevant Data Objects from multiple sources. Each Data Object also has its own dedicated page that contains information about the taxon (or taxa) the Data Object is associated with, license information, all available source and attribution information, a tool for rating the Data Object, links to other versions of the Data Object, comments on the Data Object, and, for non-text objects, a text description (caption) if available. These Data Object Pages are accessible through links from EOL Taxon Pages and through their own unique URLs (e.g. http://eol.org/data_objects/21942847). EOL curators have access to tools on the Data Object Page to control visibility and trusted status, and on image Data Object Pages, tools to crop images to create versions of thumbnail images that are shown throughout EOL. While curators can hide a Data Object or indicate its trusted status, the content itself can only be changed or updated by the provider.
Darwin Core Archive support for content ingestion
Initially, EOL harvested resource documents formatted according to an XML transfer schema drawing from standards such as Dublin Core, Darwin Core (
Most EOL content is aggregated via content partner tools (designed for projects that have large amounts of content to share) or added directly to the web site by users. Any EOL member can add and manage an EOL content partner account through their member profile (see http://www.eol.org/cp_getting_started). After supplying basic information about their project, users register one or more resource documents that contain the information they want to share. Resource documents may be customized exports from a database, they may be created by programs that parse web pages or call web services, or they may be manually assembled spreadsheets. Some resources are the result of newly published marked-up taxonomic treatments (
Currently, EOL members can add text objects, also known as articles, directly to EOL using the “Add an Article” button on the Details tab. Multimedia objects cannot be uploaded directly to EOL but must be added through partners such as Flickr, Wikimedia Commons, iNaturalist, Vimeo, YouTube, and Soundcloud.
EOL has developed a Richness Score for taxon pages (Fig.
The EOL richness score is the sum of three weighted components: breadth, depth, and diversity. Breadth considers the different media types of information objects (including the number of different subjects available for text), depth considers both average and total number of words in text objects, and diversity considers the number of different sources of information, or providers. Normalized object values are scaled to be between 0 and 1 and put on a log-based scale such that the first objects counts more than the second up to a chosen limit at which point the value is 1 and additional objects of that type have no impact on the richness. The final score is multiplied by 100, so that it ranges from 0 to 100. For more detailed information, see http://eol.org/info/richness_score.
EOL Communities provide a way to group users. The primary value of this feature at the moment is to share the management of different EOL Collections. They also provide a simple forum through the associated newsfeed. Collections provide a way for users to organize, annotate, and share the content on the site. Collections may range from species lists for local areas (e.g. Florida Native Plants) to lists of homonyms (Homonyms on EOL) to content collections for education or entertainment (e.g., X-ray Vision: Fish Inside Out). Many different types of items within EOL can be collected including Taxon, Image, Article, User, Community and even other Collection pages (e.g., a collection of video collections). Collections can be viewed as a visual gallery, a simple list, or an annotated list and can be sorted in a variety of ways including by Richness Score. Annotation fields allow Collection managers to provide notes, references, or sort fields for each item in the collection. By default, an EOL Collection is managed by the user who creates it. However, any manager can share management privileges with other EOL members or communities.
EOL provides curation tools for volunteer data curators. All curators must register under their real names. To facilitate participation of EOL members with different levels of expertise, three different curator levels are distinguished. As of April 2014, almost 300 EOL members have registered as assistant curators and over 1,300 members have been approved as full or master curators.
The Assistant Curator status requires no qualifications and conveys limited curation powers. Assistant Curators can add taxon associations to data objects (e.g., to identify organisms shown in an image), but these associations are marked as "unreviewed" until confirmed by a Full Curator. Assistant Curators can also add common names, select preferred common names, select exemplar images and articles, and crop image thumbnails. They are encouraged to add text and help find problems that Full Curators can resolve. Full Curators must have credentials (e.g. relevant professional affiliations, publications, membership in a professional association). In addition to the powers of Assistant Curators, they can trust or untrust text or multimedia objects and select preferred classifications for taxon pages. Master Curators can manage taxon concepts (overriding the automated reconciliation process by merging or splitting classifications featured on a given taxon page) and delete comments that do not adhere to EOL community policies.
Untrusted content is hidden from public view but still visible to Full and Master Curators for further review. Curation actions and comments are reported to content providers (Feedback, in Fig.
The EOL website search is configured to find scientific names and common names, with preference in search result ordering given to preferred scientific names (names that have been manually selected by curators as “preferred” for a taxon) first, followed by preferred common names, and synonym. EOL search also indexes Communities, Collections, EOL members, Data Objects, and EOL documentation pages, and search results can be filtered by these categories. If there is a best result, the system takes the user directly to that taxon page, with an option to return to the search results page to view other results.
The EOL Application Programming Interface (API) allows content indexed by EOL to be easily accessible to other websites and software developers. Through the API, applications can search EOL Taxon Pages, fetch page metadata such as names, images and text, and access hierarchy and collection information. The latest versions of API methods allow data to be returned in either XML format or the simpler JSON format. Method documentation has been improved and internationalized, and now includes forms to help users test the methods and their various parameters by interactively showing the responses. An example of a website using the EOL API to feature EOL data within their own site is the Smithsonian National Museum of Natural History’s Species of the Day widget. This widget is created using the API to draw data from a custom EOL Collection. Other examples include various games, visualizations, and other sites that re-use EOL content.
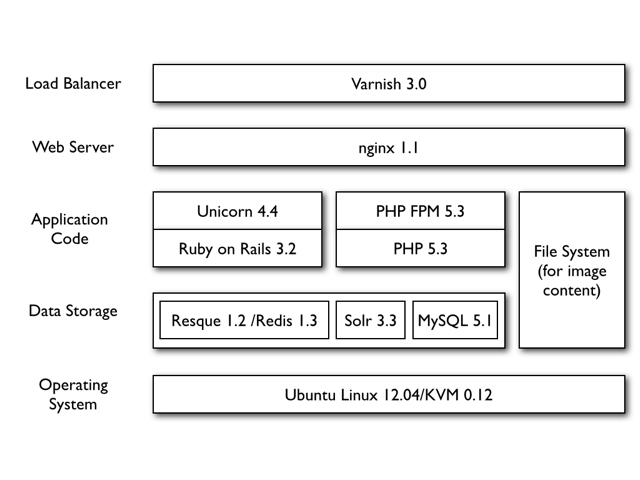
EOL Version 2 provided an opportunity to significantly improve the hardware and software infrastructure of EOL. The entire software and hardware stack supporting the serving of eol.org moved to the Research Computing group at Harvard University while remaining managed by the EOL Operations team at the Marine Biological Laboratory. The new architecture introduced KVM-based virtual machines to the infrastructure, allowing a more efficient use of resources and faster deployment of new infrastructure services to support the hosting of the site (Fig.
The EOL technical team uses a modified version of the Scrum software development framework (
Audience
EOL has a worldwide audience including experts, enthusiasts and casual visitors. About 39% of user sessions originate in the United States and more than 47% of user sessions originate in countries where English is not an official language. Starting with v2, visitors registering to become EOL members were invited to select one or more audience categories to describe themselves. Of 6,410 people who self-identified by 18 April 2014, 47% chose "enthusiast", 36% chose "student", 20% chose "educator", 18% chose "citizen scientist", and 20% chose "professional scientist". However, this distribution may not reflect the more than 73,000 current EOL members or the vastly larger number of visitors who never register or who encounter EOL content primarily via social media channels.
Experts and enthusiasts are encouraged to participate in EOL as content curators. As of April 2014, almost 300 EOL members have registered as assistant curators and over 1,300 members have been approved as full or master curators.
At least in North America, the formal education audience is an important demographic for EOL. We see from Google Analytics that there are increases in the use of the site when most schools are in session. The EOL Learning & Education group also actively posts information on about 15 listservs, including the National Science Teachers Association (NSTA), Scuttlebutt (NOAA Marine Education site) and the Ecological Society of America's EcoLogic Listserv.
Additional information
Milestones
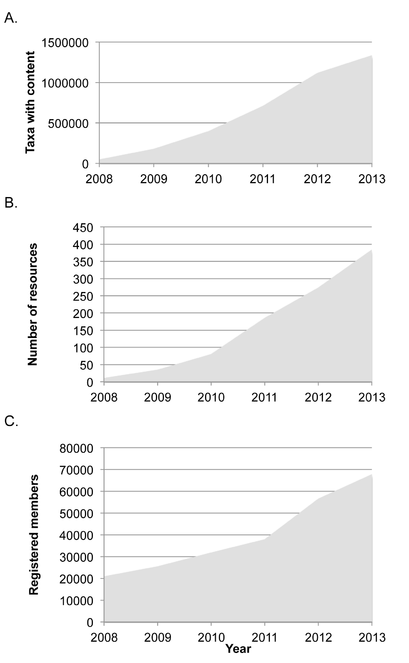
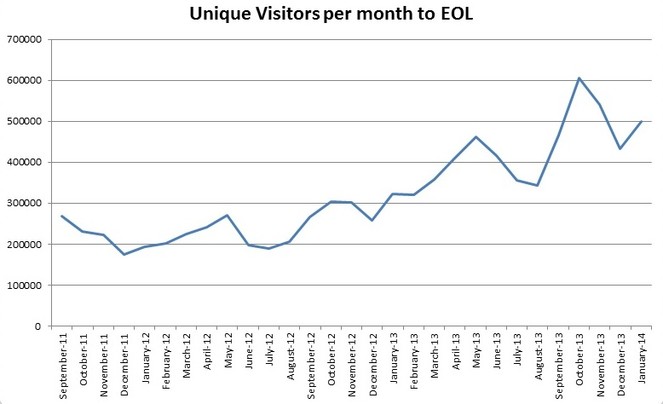
EOL’s growth in overall information, provider resources, and membership has steadily increased (Fig.
Growth in Encyclopedia of Life from 2008-2013. A. Taxon pages with content (at least one text article, image, map, video, or sound) (Suppl. material
Still, most EOL pages remain without content, i.e., EOL provides nothing but a taxon name, and in some cases author information and a reference. Overall, EOL has indexed about 3.5 million taxa. This represents most of the 1.9 million extant (
Closer examination indicates that EOL has an uneven distribution of content across languages, licenses, and topics. While EOL has vernacular names in 163 languages (Table
| Language | Common Names |
|---|---|
| English | 690163 |
| Spanish | 114579 |
| Chinese | 87643 |
| French | 85973 |
| German | 69945 |
| Japanese | 51432 |
| Portuguese | 42497 |
| Italian | 39264 |
| Czech | 37455 |
| Russian | 35379 |
| Danish | 30775 |
| Dutch | 30775 |
| Finnish | 29785 |
| Polish | 24918 |
| Other | 280057 |
| Language | Articles |
|---|---|
| English | 3096313 |
| Spanish | 58978 |
| Chinese | 11678 |
| Arabic | 4807 |
| Portuguese | 2373 |
| Dutch | 1143 |
| Indonesian | 173 |
| French | 107 |
| Other | 10180 |
Subjects of text articles. Combined topics include Wikipedia (n = 223571), Description (n = 49074), General Description (n = 45887), Brief Summary (n = 26862) and Biology (n = 8929). Subjects with fewer than 100 articles are not shown (Procedures, Legislation, Identification Resources, Systematics or Phylogenetics, Development).
| Subject | Articles |
|---|---|
| Distribution | 805503 |
| Molecular Biology | 434545 |
| Combined Topics | 354322 |
| Type Information | 326720 |
| Habitat | 292478 |
| Conservation Status | 144969 |
| Threats | 94140 |
| Morphology | 66571 |
| Conservation | 65618 |
| Diagnostic Description | 61512 |
| Management | 57894 |
| Trends | 57888 |
| Size | 55453 |
| Description | 49074 |
| Associations | 38677 |
| Taxon Biology | 26861 |
| Uses | 24458 |
| Trophic Strategy | 21563 |
| Population Biology | 17767 |
| Taxonomy | 16301 |
| Ecology | 15060 |
| Reproduction | 14996 |
| Notes | 14440 |
| Migration | 13991 |
| Cyclicity | 11880 |
| Life Cycle | 9759 |
| Life Expectancy | 8875 |
| Behavior | 6391 |
| Key | 6118 |
| Diseases | 4325 |
| Use | 4283 |
| Evolution | 2158 |
| Risk Statement | 2022 |
| Look Alikes | 1897 |
| Dispersal | 1649 |
| Functional Adaptations | 1438 |
| Genetics | 1000 |
| Growth | 785 |
| Barcode | 720 |
| Education Resources | 646 |
| Physiology | 269 |
| Cytology | 129 |
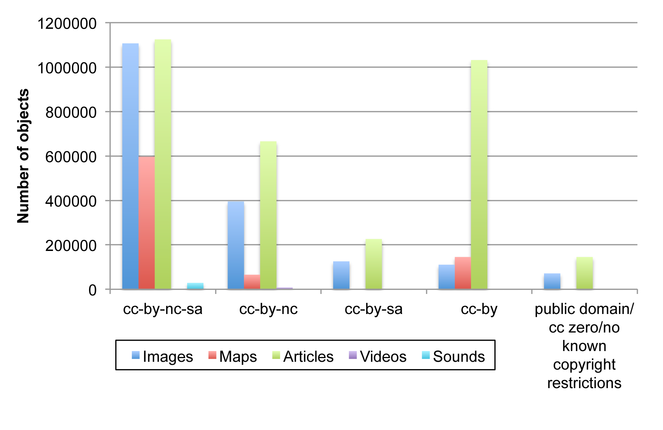
Distribution of Creative Commons and other licenses for data objects on EOL. CC-BY = Creative Commons Attribution license; NC = Non-commercial restriction; SA = Share-alike restrictions. Objects with gnu-gpl/gnu-fdl licenses (3903 images and 21 text articles) are not shown. Overall, as of July 2013, EOL has 3,192,609 text articles, 1,812,295 images, 806,664 maps, 30,366 sounds, and 10,219 videos (Suppl. material
To date, users have created more than 5,000 EOL Collections. Many collections (approximately 35%) are for specific geographic regions and represent user-generated checklists that could be useful for refining map queries in areas where occurrence data are not yet available. Presence of a taxon or object in many user-generated Collections could be used (by EOL or by others) to sort or filter search results so that they are most relevant to user needs. Collection statistics, along with traffic statistics, could also help researchers explore the factors that make an organism or data object more engaging to broad audiences.
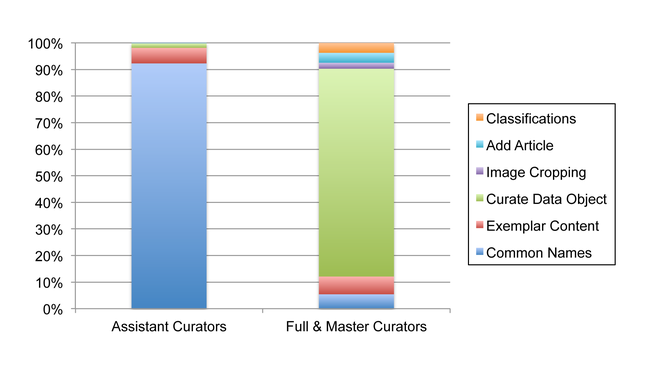
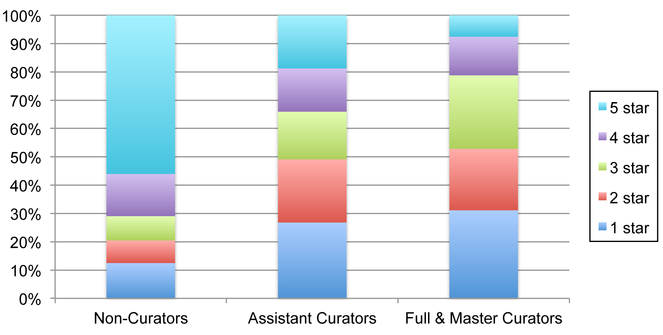
Though there is room for growth in curation activity, EOL is increasingly in a position to improve data quality across its network of providers. In July 2013, EOL had 1,258 registered curators (250 Assistant, 1,001 Full, 7 Master) of which 163 have been active in the last 12 months. In comparison, iNaturalist has 94 curators and the World Register of Marine Species has 826 editors (a thoughtful analysis of curation power across projects with different models is beyond the scope of this paper). The majority of data objects are considered trusted (92%), most having been acquired from authoritative sources. An average of 905 objects per month are being curated. Assistant and Full Curators have different patterns of activity, not surprisingly given their different access to tools (Fig.
Activity patterns of EOL Assistant Curators compared to Full and Master Curators. Only Full and Master Curators can select preferred classifications and change the visibility and trust status of text and multimedia objects. Data Object curation by Assistant Curators is limited to adding associations between Data Objects and taxa.
In the period from August 2012 through July 2013, EOL was visited by 3.7 million unique users. About 44% of visits are from North America (including Mexico). Thirteen countries on other continents contributed a significant number of visits.
Discussion
EOL has established its role of improving access to biodiversity information by aggregating and standardizing descriptive information and multimedia objects currently available across many otherwise isolated resources. It provides the infrastructure to connect both major hubs and independent projects (
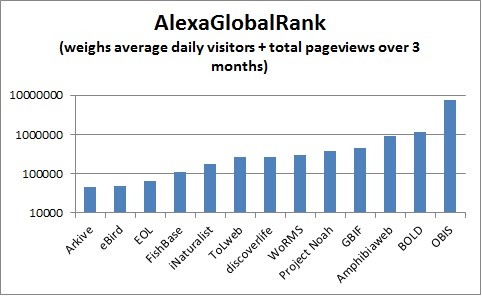
Global Rank for biodiversity web sites per http://www.alexa.com/, 02/26/2014. The rank is calculated using a combination of average daily visitors to this site and pageviews on this site over the past 3 months. Lower numbers indicate greater importance, as the site with the highest combination of visitors and pageviews is ranked #1. Note however that Alexa rankings are known to be subject to considerable sampling bias since they are largely based on the behavior of users browsing with an Alexa-compatible toolbar (
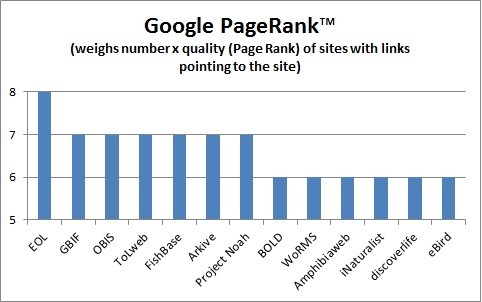
Google PageRank™ of various biodiversity websites, per http://www.prchecker.info/, 02/22/2014. Larger numbers indicate greater importance, and webpages with a higher PageRank are more likely to appear at the top of Google search results.
EOL complements long-term archives and metadata registries, e.g. DataONE (
By taking a phased approach (phase 1 of core infrastructure and phase 2 of engagement), EOL has successfully built a professional, usable platform at a scale appropriate to its task of serving global biological information to multiple international audiences. Because it is scalable, as EOL grows, its Richness Scores can be used to assess the availability and quality of knowledge across the tree of life, especially when extended to structured data. The scores could also enable assessment of individual contributions and standardization (
Several challenges remain to be tackled in future phases. While there is some evidence (growth in collections, emergence of third party applications, curator activity, user traffic) of effective impact on and engagement by various audiences, tools for community and curator engagement are not as successful as hoped and so they may require more tailored experiences and effective feedback (
The next phase of EOL moves beyond the limits of encyclopedic text and multimedia to add the ability to ingest and serve highly structured data (numeric and controlled vocabulary terms with rich semantics) about the attributes of and relationships among organisms (Parr et al. in review). In the same way that EOL has helped to bring together and connect text and media from isolated sources, we aggregate structured data to provide a broad-scale view of analyzable biodiversity data. EOL’s standardized open access also facilitates new text mining or crowd-sourcing efforts to extract structured data about biological diversity, e.g.
Acknowledgements
Support was provided by John D. and Catherine T. MacArthur Foundation, Alfred P. Sloan Foundation, Smithsonian Institution, Marine Biological Laboratory, and Harvard University. The production hardware infrastructure for the EOL website is supported by the Harvard Faculty of Arts and Sciences (FAS) Sciences Division Research Computing Group. We thank all of our providers and global partners, Eli Agbayani, Tracy Barbaro, Dana Campbell, Vitthal Kudal, Erick Mata, David Patterson, and Mark Westneat. Leo Shapiro and Dawn Field provided helpful comments on the manuscript.
Author contributions
Conceived and designed the experiments: CSP NW BC MS. Coded the software: PL JR LW AG. Managed data ingestion: JAH, KSS CSP PL. Provided detailed requirements and tested the software: JAH KSS CSP JTGH MS. Performed analyses: KSS CSP. Wrote the paper: CSP NW JAH KSS KL PL LW AG.
References
- Visually Exploring Social Participation in Encyclopedia of Life.Social Informatics,Lausanne,Dec 14-16, 2012.8pp. https://doi.org/10.1109/socialinformatics.2012.51
- How many named species are valid?Proceedings of the National Academy of Sciences (USA)99(6):3706‑3711.
- Linking multiple biodiversity informatics platforms with Darwin Core Archives.Biodiversity Data Journal2:e1039. https://doi.org/10.3897/bdj.2.e1039
- GenBank.Nucleic Acids Research41:D36‑D42. https://doi.org/10.1093/nar/gks1195
- Biodiversity update — Progress in taxonomy.Himalayan Journal of Sciences1(2):83‑84. https://doi.org/10.3126/hjs.v1i2.202
- The taxonomic name resolution service: an online tool for automated standardization of plant names.BMC Bioinformatics14(1):16. https://doi.org/10.1186/1471-2105-14-16
- Progressive Enhancement and the Future of Web Design.REPRINTED FROM WEBMONKEY,1pp. URL: http://www.hesketh.com/thought-leadership/our-publications/progressive-enhancement-and-future-web-design
- Numbers of living species in Australia and the World Report. Commonwealth of Australia.Department of the Environment and Water Resources,84pp. URL: http://www.environment.gov.au/biodiversity/abrs/publications/other/species-numbers/index.html
- Towards a data publishing framework for primary biodiversity data: challenges and potentials for the biodiversity informatics community.BMC Bioinformatics10:S2. https://doi.org/10.1186/1471-2105-10-s14-s2
- Global coordination and standardisation in marine biodiversity through the World Register of Marine Species (WoRMS) and related databases.PloS one8(1):e51629. https://doi.org/10.1371/journal.pone.0051629
- What is EOL? - Information and pictures of all species known to science.Encyclopedia of Life,1pp. URL: http://eol.org/about
- Content assessment of the primary biodiversity data published through GBIF network: Status, Challenges and Potentials.Biodiversity Informatics8(2):94‑172.
- Data hosting infrastructure for primary biodiversity data.BMC Bioinformatics12:S5. https://doi.org/10.1186/1471-2105-12-s15-s5
- Creative Commons licenses and the non-commercial condition: Implications for the re-use of biodiversity information.ZooKeys150:127‑149. https://doi.org/10.3897/zookeys.150.2189
- How many species of flowering plants are there?Proceedings of the Royal Society B: Biological Sciences278(1705):554‑559. https://doi.org/10.1098/rspb.2010.1004
- Why impact factors don't work for taxonomy.Nature415(6875):957‑957. https://doi.org/10.1038/415957a
- Organizing our knowledge of biodiversity.Bulletin of the American Society for Information Science and Technology37(4):38‑42. https://doi.org/10.1002/bult.2011.1720370411
- The Metadata Coverage Index (MCI): A standardized metric for quantifying database metadata richness.Standards in Genomic Sciences6(3):444‑453. https://doi.org/10.4056/sigs.2675953
- How reliable are website rankings? Implications for e-business advertising and internet search.Issues in Information Systems7(2):233‑238.
- The Tree of Life Web Project.Zootaxa40:19‑40. URL: http://tolweb.org
- Ramping up biodiversity discovery via online quantum contributions.Trends in Ecology & Evolution27(2):72‑77. https://doi.org/10.1016/j.tree.2011.10.010
- Biology Needs a Modern Assessment System for Professional Productivity.BioScience61(8):619‑625. https://doi.org/10.1525/bio.2011.61.8.8
- Participatory design of DataONE—Enabling cyberinfrastructure for the biological and environmental sciences.Ecological Informatics11:5‑15. https://doi.org/10.1016/j.ecoinf.2011.08.007
- From taxonomic literature to cybertaxonomic content.BMC Biology10(1):87. https://doi.org/10.1186/1741-7007-10-87
- The animal diversity web.University of Michigan,5286pp. URL: http://animaldiversity.org
- The Encyclopedia of Life, Biodiversity Heritage Library, Biodiversity Informatics and Beyond Web 2.0.First Monday13(8):2011‑2013. https://doi.org/10.5210/fm.v13i8.2226
- The PageRank Citation Ranking: Bringing Order to the Web.World Wide Web Internet And Web Information Systems54:1‑17. URL: http://ilpubs.stanford.edu:8090/422
- bioGUID: resolving, discovering, and minting identifiers for biodiversity informatics.BMC Bioinformatics10:S5. https://doi.org/10.1186/1471-2105-10-s14-s5
- Evolutionary informatics: unifying knowledge about the diversity of life.Trends in Ecology & Evolution27(2):94‑103. https://doi.org/10.1016/j.tree.2011.11.001
- Visualizations for taxonomic and phylogenetic trees.Bioinformatics20(17):2997‑3004. https://doi.org/10.1093/bioinformatics/bth345
- Principles for a names-based cyberinfrastructure to serve all of biology.Zootaxa1950:153‑163.
- Scientific names of organisms: attribution, rights, and licensing.BMC Research Notes7(1):79. https://doi.org/10.1186/1756-0500-7-79
- The Measurement of Species Diversity.Annual Review of Ecology and Systematics5(1):285‑307. https://doi.org/10.1146/annurev.es.05.110174.001441
- Semantic tagging of and semantic enhancements to systematics papers: ZooKeys working examples.ZooKeys50:1‑16. https://doi.org/10.3897/zookeys.50.538
- Bringing Fossils to Life: An Introduction to Paleobiology.3rd Edition.Columbia University Press,New York,672pp.
- Species 2000 & ITIS Catalogue of Life.1,352,112pp. URL: www.catalogueoflife.org/col/
- Supporting content curation communities: The case of the Encyclopedia of Life.Journal of the American Society for Information Science and Technology63(6):1092‑1107. https://doi.org/10.1002/asi.22633
- Dynamic changes in motivation in collaborative citizen-science projects.Computer Supported Cooperative Work,Bellevue, WA,Feb 11-15, 2012.10pp. https://doi.org/10.1145/2145204.2145238
- Managing Biodiversity Knowledge in the Encyclopedia of Life.BNCOD 2008 Biodiversity Informatics Workshop,Cardiff University,10th July 2008.2pp. URL: http://biodiversity.cs.cf.ac.uk/bncod/SchopfEtAl.pdf
- Agile project management with Scrum. Van Steenburgh R, Engelman L, Atkins K (Eds).Microsoft Press,155pp. URL: http://www.bjla.dk/VideregUdvikling/DM052/ScrumProjectManagementPart00.pdf
- Scratchpads 2.0: a Virtual Research Environment supporting scholarly collaboration, communication and data publication in biodiversity science.Zookeys2011(150):53‑70. https://doi.org/10.3897/zookeys.150.2193
- Stork NE (1997) Measuring global diversity and its decline. In: Reaka-Kudla ML, Wilson DE, Wilson EO (Eds) Biodiversity II: Understanding and Protecting Our Biological Resources.Joseph Henry Press,Washington, DC,pp. 41–68pp.
- Web Content Accessibility Guidelines 2.0.World Wide Web Internet And Web Information Systems27:1171‑1172. URL: http://www.w3.org/TR/WCAG20/
- Knowledge Extraction and Semantic Annotation of Text from the Encyclopedia of Life.PLoS ONE9(3):e89550. https://doi.org/10.1371/journal.pone.0089550
- The Dryad Digital Repository: Published evolutionary data as a part of the greater data ecosystem.Nature Precedings1(4595):1. URL: http://hdl.handle.net/10101/npre.2010.4595.1
- Name Matters: Taxonomic Name Recognition (TNR) in Biodiversity Heritage Library (BHL).iConference 2010 Proceedings. University of Illinois.2010:3‑7. URL: http://hdl.handle.net/2142/14919
- Mapping the biosphere: exploring species to understand the origin, organization and sustainability of biodiversity.Systematics and biodiversity10:1‑20. https://doi.org/10.1080/14772000.2012.665095
- Darwin Core: an evolving community-developed biodiversity data standard.PLoS ONE7(1):e29715. https://doi.org/10.1371/journal.pone.0029715
- The encyclopedia of life.Trends Ecol Evol18:77‑80.
- Yoon N, Rose J (2001) An Information Model for the Representation of Multiple Biological Classifications. Lecture Notes in Computer Science — ICCS International Conference San Francisco, CA, USA, May 28–30, 2001 Proceedings, Part I.2073.Springer Berlin Heidelberghttps://doi.org/10.1007/3-540-45545-0_106
Supplementary materials
Taxon pages with content (at least one text article, image, map, video, or sound).
Published resources (content import files). A provider may submit more than one resource file, for example when providing different kinds of content.
Registered EOL members.
Distribution of Creative Commons and other licenses for data objects on EOL.
Activity patterns of EOL Assistant Curators compared to Full and Master Curators.
Data Object rating patterns of EOL members in relation to their curator status.