|
Biodiversity Data Journal :
Research Article
|
|
Corresponding author: Matthew L. Bowser (matt_bowser@fws.gov)
Academic editor: Pavel Stoev
Received: 13 Oct 2016 | Accepted: 04 Jan 2017 | Published: 12 Jan 2017
© 2017 Matthew Bowser, John Morton, John Hanson, Dawn Magness, Mallory Okuly
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Bowser M, Morton J, Hanson J, Magness D, Okuly M (2017) Arthropod and oligochaete assemblages from grasslands of the southern Kenai Peninsula, Alaska. Biodiversity Data Journal 5: e10792. https://doi.org/10.3897/BDJ.5.e10792
|
|
Abstract
Background
By the end of this century, the potential climate-biome of the southern Kenai Peninsula is forecasted to change from transitional boreal forest to prairie and grasslands, a scenario that may already be playing out in the Caribou Hills region. Here, spruce (Picea × lutzii Little [glauca × sitchensis]) forests were heavily thinned by an outbreak of the spruce bark beetle (Dendroctonus rufipennis (Kirby, 1837)) and replaced by the native but invasive grass species, Calamagrostis canadensis (Michx.) P. Beauv. As part of a project designed to delimit and characterize potentially expanding grasslands in this region, we sought to characterize the arthropod and earthworm communities of these grasslands.
We also used this sampling effort as a trial of applying high-throughput sequencing metabarcoding methods to a real-world inventory of terrestrial arthropods.
New information
We documented 131 occurrences of 67 native arthropod species at ten sites, characterizing the arthropod fauna of these grasslands as being dominated by Hemiptera (60% of total reads) and Diptera (38% of total reads). We found a single exotic earthworm species, Dendrobaena octaedra (Savigny, 1826), at 30% of sites and one unidentified enchytraeid at a single site. The utility of high-throughput sequencing metabarcoding as a tool for bioassessment of terrestrial arthropod assemblages was confirmed.
Keywords
metagenomics, Arthropoda, exotic earthworms, Illumina MiSeq
Introduction
Background
By the end of this century, the potential climate-biome of the southern Kenai Peninsula is forecasted to change from transitional boreal forest to prairie and grasslands (
Following this outbreak, the native but invasive herbaceous species Calamagrostis canadensis (Michx.) P. Beauv. and Chamerion angustifolium (L.) Holub increased in abundance (
Of the fauna of northern grasslands, arthropods are among the most abundant, diverse, and ecologically important (see
With the exception of
Lumbricid earthworms are relatively recent arrivals to Alaska translocated from the Palearctic by human activities (
Metabarcoding as a tool for assessing terrestrial arthropod assemblages
The investigator seeking to characterize assemblages of arthropods or of other diverse groups is currently presented with a wide and growing range of options for obtaining species identifications including traditional, specimen-based, morphological identifications; Sanger sequencing of individual specimens using DNA barcodes (
High-throughput sequencing metabarcoding methods have been advocated for biomonitoring of arthropod communities because they have the potential to be quick and comparatively inexpensive (
Obtaining correct species identifications from HTS methods requires a well-curated library of sequences from identified specimens (
In this small project we applied HTS metabarcoding methods to a real-world inventory with a vision of applying similar methods to future biomonitoring efforts.
Materials and Methods
Study area and study design
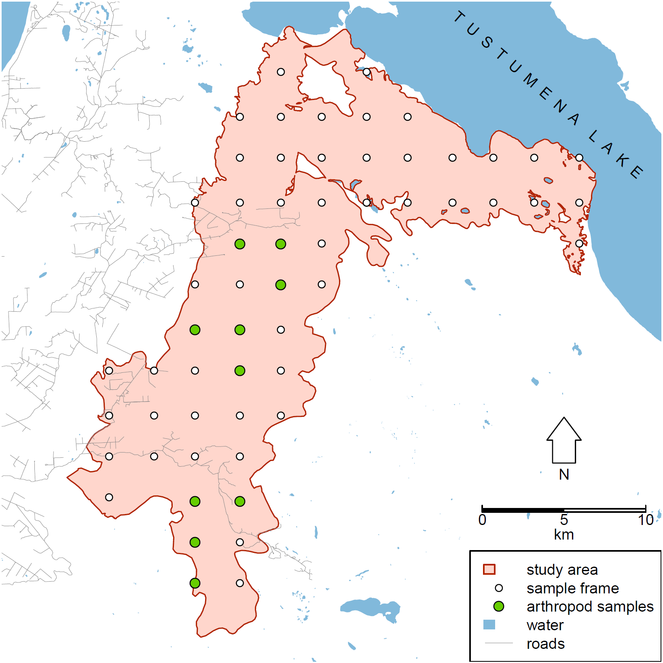
Our study area was a 37,790 ha union of major fire polygons south of Tustumena Lake on the southern Kenai Peninsula. This included the 1994 Windy Point Fire, 1996 Crooked Creek Fire, 2005 Fox Creek Fire, and 2007 Caribou Hills Fire.
Within this area, we chose as a sample frame to use centroids of the 250 m pixels from the Alaska eMODIS product (
Field methods
Sampling sites were accessed using a Bell 206B Jetranger on July 18-19, 2015. Only when a site was determined from the air to be a non-wetland grassland as defined by
All plant species within a 5.64 m radius, 100 m2 circular plot centered on the plot coordinates were recorded. Plants that could not be identified in the field were collected.
Earthworms were collected at each plot using methods similar to those of
At ten sites we collected a single sample of arthropods by sweeping the same 5.64 m radius plot in under five minutes using a BioQuip™ model 7112CP net with 30.5 cm diameter, approximately 24 × 20 per inch mesh BioQuip™ model 7112CPA net bag and a BioQuip™ model 7312AA 30.5 cm extension handle. Sweep net samples were placed in 250 ml Nalgene® vials filled with Uni-Gard -100 propylene glycol antifreeze, then stored in a -23°C freezer.
Laboratory methods
Plant specimens were identified in the laboratory using the keys of
We identified earthworm specimens visually using the key of
Arthropods were separated from vegetation and debris by hand under a dissecting microscope. At the same time, all athropods were tallied and coarsely identified, generally to orders but sometimes to families, genera, and species that could be quickly identified by sight. We made no attempt to account for the varying sizes of different arthropods.
Specimen data (Table
Sample collection data. Complete collection data including photographs of the sampling sites are available from Arctos. Dates are given in ISO 8601 format.
|
Arctos GUID |
BioSample |
latitude |
longitude |
date |
|
|
|
2015-07-19 |
||
|
|
|
2015-07-19 |
||
|
|
|
2015-07-19 |
||
|
|
|
2015-07-18 |
||
|
|
|
2015-07-18 |
||
|
|
|
2015-07-18 |
||
|
|
|
2015-07-18 |
||
|
|
|
2015-07-18 |
||
|
|
|
2015-07-19 |
||
|
|
|
2015-07-19 |
Samples were shipped in propylene glycol to RTL Genomics in Lubbock, Texas for sequencing. Upon arrival, samples were removed from propylene glycol and rinsed with 100% ethanol. Ethanol rinse was decanted and enough 100% ethanol was added to the container to cover the arthropods. Samples were stored in Ethanol for 21 days. Samples were then rinsed in PBS, then 400 μl of PBS was added to the sample and the sample was ground using an Omni Tissue Homogenizer. Extraction was performed using MoBioPower soil extraction kit with an overnight incubation at 37°C. To elute the sample 50 μl of prewarmed elution buffer was added to the column membrane and incubated at room temperature for 2 min, then spun down. The elutate was place back on the column and incubated another 2 min, then spun down.
We used the forward primer mlCOIintF (GGWACWGGWTGAACWGTWTAYCCYCC) and reverse primer HCO2198 (TAAACTTCAGGGTGACCAAAAAATCA). These primers used previously by
Sequence data were submitted to GenBank's Sequence Read Archive (BioProject: PRJNA321553).
Total molecular lab processing cost was $1,115 ($111.50 per sample) and sequencing results were delivered 68 days after samples had been received by RTL Genomics.
Library construction and metagenomic analysis
For the present study, we constructed an Alaska vicinity reference library by downloading publicly available COI data from BOLD on January 20-21, 2016, entering the search terms "Arthropoda[tax] Alaska[geo]" and similarly structured searches for arthropod sequences from the Yukon Territory, British Columbia, Chukot Autonomous Okrug, and Kamchatka Krai, yielding an initial library of 236,830 records including 6,677 unique species name strings.
A metagenomic analysis was performed using the cloud-based Galaxy platform (
Where one of a pair of reads had a read length less than 250 bp, these were filtered out using R version 3.2.2 (
Identifications were improved iteratively. First, initial identifications were obtained by querying the cluster centroids against our reference library using VSearch search version 1.9.7.0 (
For all library records that were matched by our queries and that lacked species names we added identifications by submitting them to BOLD's Identification Request service and updating our library records with any identification improvements. In cases where no species names were available, we constructed provisional names incorporating BOLD BIN URIs (
For cluster centroids that were not matched by our library, we queried these against the BOLD database using the bold() function from the bold package for R, version 0.3.5 (
Library composition by country and province/state. Country and province values of NA indicate sequences lacking corresponding geographic data.
|
Country |
Province |
Number of records |
|
Canada |
Alberta |
2 |
|
Canada |
British Columbia |
193,410 |
|
Canada |
Manitoba |
4 |
|
Canada |
Newfoundland and Labrador |
2 |
|
Canada |
Northwest Territories |
1 |
|
Canada |
Ontario |
2 |
|
Canada |
Prince Edward Island |
2 |
|
Canada |
Quebec |
3 |
|
Canada |
Yukon Territory |
35,406 |
|
Russia |
Chukot Autonomous Okrug |
406 |
|
Russia |
Kamchatka Krai |
665 |
|
United States |
Alaska |
6,923 |
|
NA |
NA |
11 |
|
Total |
236,837 |
Composition of sweep net samples as determined by sight identifications.
|
Class |
Order |
Family |
Genus |
Species |
Quantity |
|
Arachnida |
Acari |
1 |
|||
|
Total Acari |
1 |
||||
|
Arachnida |
Araneae |
2 |
|||
|
Arachnida |
Araneae |
Tetragnathidae |
Tetragnatha |
2 |
|
|
Arachnida |
Araneae |
Thomisidae |
Misumena |
Misumena vatia (Clerck, 1757) |
1 |
|
Total Araneae |
5 |
||||
|
Insecta |
Coleoptera |
6 |
|||
|
Insecta |
Coleoptera |
Elateridae |
1 |
||
|
Insecta |
Coleoptera |
Staphylinidae |
2 |
||
|
Total Coleoptera |
9 |
||||
|
Insecta |
Diptera |
135 |
|||
|
Insecta |
Diptera |
Agromyzidae |
2 |
||
|
Insecta |
Diptera |
Bibionidae |
1 |
||
|
Insecta |
Diptera |
Chironomidae |
23 |
||
|
Insecta |
Diptera |
Culicidae |
3 |
||
|
Insecta |
Diptera |
Empididae |
45 |
||
|
Insecta |
Diptera |
Ephydridae |
1 |
||
|
Insecta |
Diptera |
Lauxaniidae |
Lauxania |
Lauxania shewelli Perusse & Wheeler, 2000 |
14 |
|
Insecta |
Diptera |
Phoridae |
30 |
||
|
Insecta |
Diptera |
Pipunculidae |
2 |
||
|
Insecta |
Diptera |
Rhagionidae |
Symphoromyia |
5 |
|
|
Insecta |
Diptera |
Scathophagidae |
4 |
||
|
Insecta |
Diptera |
Simuliidae |
13 |
||
|
Insecta |
Diptera |
Sphaeroceridae |
2 |
||
|
Insecta |
Diptera |
Syrphidae |
1 |
||
|
Total Diptera |
281 |
||||
|
Insecta |
Hemiptera |
238 |
|||
|
Insecta |
Hemiptera |
Aphididae |
39 |
||
|
Insecta |
Hemiptera |
Cicadellidae |
257 |
||
|
Insecta |
Hemiptera |
Miridae |
29 |
||
|
Insecta |
Hemiptera |
Miridae |
Irbisia |
28 |
|
|
Insecta |
Hemiptera |
Nabidae |
Nabis |
9 |
|
|
Insecta |
Hemiptera |
Psyllidae |
80 |
||
|
Total Hemiptera |
680 |
||||
|
Insecta |
Hymenoptera |
27 |
|||
|
Insecta |
Hymenoptera |
Ichneumonidae |
9 |
||
|
Insecta |
Hymenoptera |
Sphecidae |
2 |
||
|
Insecta |
Hymenoptera |
Tenthredinidae |
6 |
||
|
Insecta |
Hymenoptera |
Torymidae |
Torymus |
2 |
|
|
Total Hymenoptera |
46 |
||||
|
Insecta |
Lepidoptera |
3 |
|||
|
Total Lepidoptera |
3 |
||||
|
Insecta |
Psocoptera |
4 |
|||
|
Total Psocoptera |
4 |
Summary of occurrence data from the metagenomic analysis. BIN: BOLD Barcode Index Numbers from matched sequences. f: frequency of occurrence, the proportion of all samples in which each taxonomic unit was detected.
|
Order |
Family |
Species |
BIN |
f |
|
Araneae |
Thomisidae |
Misumena vatia |
0.1 |
|
|
Coleoptera |
Chrysomelidae |
Altica tombacina |
0.2 |
|
|
Coleoptera |
Elateridae |
Hypnoidus bicolor |
0.1 |
|
|
Diptera |
Anthomyiidae |
Anthomyiidae sp. BOLD:AAG2469 |
0.3 |
|
|
Diptera |
Anthomyiidae |
Botanophila relativa |
0.1 |
|
|
Diptera |
Anthomyiidae |
Botanophila rubrigena |
0.1 |
|
|
Diptera |
Anthomyiidae |
Delia echinata |
0.1 |
|
|
Diptera |
Anthomyiidae |
Hylemya variata |
0.4 |
|
|
Diptera |
Anthomyiidae |
Paradelia brunneonigra |
0.1 |
|
|
Diptera |
Anthomyiidae |
Pegomya sp. BOLD:AAG2506 |
0.1 |
|
|
Diptera |
Anthomyzidae |
Anthomyza sp. BOLD:AAL8100 |
0.2 |
|
|
Diptera |
Bibionidae |
Bibionidae sp. BOLD:ACG6252 |
0.2 |
|
|
Diptera |
Chironomidae |
Metriocnemus sp. BOLD:ACB8808 |
0.1 |
|
|
Diptera |
Chironomidae |
Smittia sp. 16ES |
0.1 |
|
|
Diptera |
Chironomidae |
Smittia sp. ES12 |
0.1 |
|
|
Diptera |
Culicidae |
Aedes pullatus |
0.1 |
|
|
Diptera |
Empididae |
Empididae sp. BOLD:AAF9792 |
0.3 |
|
|
Diptera |
Fanniidae |
Fannia aethiops |
0.5 |
|
|
Diptera |
Fanniidae |
Fannia serena |
0.1 |
|
|
Diptera |
Heleomyzidae |
Suillia convergens |
0.1 |
|
|
Diptera |
Hybotidae |
Euthyneura sp. BOLD:AAF9859 |
0.1 |
|
|
Diptera |
Lauxaniidae |
Lauxania shewelli |
0.4 |
|
|
Diptera |
Muscidae |
Coenosia impunctata |
0.5 |
|
|
Diptera |
Muscidae |
Hydrotaea militaris |
0.3 |
|
|
Diptera |
Muscidae |
Muscidae sp. BOLD:ACL9946 |
0.1 |
|
|
Diptera |
Muscidae |
Myospila meditabunda |
0.1 |
|
|
Diptera |
Phoridae |
Megaselia diversa |
0.2 |
|
|
Diptera |
Phoridae |
Phoridae sp. BOLD:AAG3234 |
0.1 |
|
|
Diptera |
Phoridae |
Phoridae sp. BOLD:AAL9069 |
0.1 |
|
|
Diptera |
Pipunculidae |
Pipunculus campestris |
0.2 |
|
|
Diptera |
Pipunculidae |
Pipunculus hertzogi |
0.5 |
|
|
Diptera |
Pipunculidae |
Tomosvaryella sp. BOLD:AAG3766 |
0.1 |
|
|
Diptera |
Psychodidae |
Psychoda phalaenoides |
0.1 |
|
|
Diptera |
Rhagionidae |
Symphoromyia sp. BOLD:AAP6399 |
0.4 |
|
|
Diptera |
Scathophagidae |
Scathophaga furcata |
0.2 |
|
|
Diptera |
Scathophagidae |
Scathophaga suilla |
0.1 |
|
|
Diptera |
Sciaridae |
Cratyna sp. BOLD:AAP6470 |
0.1 |
|
|
Diptera |
Sciaridae |
Sciaridae sp. BOLD:AAH3999 |
0.1 |
|
|
Diptera |
Sepsidae |
Sepsis neocynipsea |
0.5 |
|
|
Diptera |
Simuliidae |
Simulium arcticum complex |
0.1 |
|
|
Diptera |
Simuliidae |
Simulium venustum complex |
0.2 |
|
|
Diptera |
Syrphidae |
Hiatomyia sp. BOLD:AAZ5940 |
0.1 |
|
|
Hemiptera |
Aphididae |
Macrosiphum euphorbiae |
0.2 |
|
|
Hemiptera |
Cicadellidae |
Balclutha sp. BOLD:AAG8963 |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Boreotettix sp. |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Diplocolenus evansi |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Empoasca luda |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Euscelis monodens sp. nov |
0.5 |
|
|
Hemiptera |
Cicadellidae |
Idiocerus sp. BOLD:ACB0208 |
0.2 |
|
|
Hemiptera |
Cicadellidae |
Latalus tatraensis |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Limotettix dasidus |
0.1 |
|
|
Hemiptera |
Cicadellidae |
Sonronius dahlbomi |
0.8 |
|
|
Hemiptera |
Cicadellidae |
Twiningia fasciata |
0.1 |
|
|
Hemiptera |
Miridae |
Irbisia sericans |
0.1 |
|
|
Hemiptera |
Miridae |
Mecomma gilvipes |
0.3 |
|
|
Hemiptera |
Miridae |
Salignus tahoensis |
0.2 |
|
|
Hemiptera |
Psyllidae |
Craspedolepta alaskensis |
0.9 |
|
|
Hemiptera |
Psyllidae |
Craspedolepta subpunctata |
0.3 |
|
|
Hymenoptera |
Braconidae |
Microgaster jft23 |
0.1 |
|
|
Hymenoptera |
Ichneumonidae |
Mesochorus prolatus |
0.1 |
|
|
Hymenoptera |
Ichneumonidae |
Orthocentrinae sp. BOLD:AAH1521 |
0.1 |
|
|
Hymenoptera |
Ichneumonidae |
Polysphincta limata |
0.1 |
|
|
Hymenoptera |
Tenthredinidae |
Amauronematus fallax |
0.1 |
|
|
Lepidoptera |
Noctuidae |
Alypia langtoni |
0.1 |
|
|
Lepidoptera |
Plutellidae |
Plutella hyperboreella |
0.1 |
|
|
Lepidoptera |
Tortricidae |
Argyrotaenia occultana |
0.1 |
|
|
Psocoptera |
Caeciliusidae |
Valenzuela flavidus |
0.1 |
The VSearch step was repeated using the improved library and the resulting occurrence data were submitted to Arctos as observation records (GUIDs: UAMObs:Ento:235609–UAMObs:Ento:235739).
We repeated the VSearch search identification step against our improved library using the same parameters. For the purpose of reporting species occurrence we exlcuded all clusters where read counts were four or less and all clusters where the VSearch search similarity values were less than 0.91. Clusters matching human COI were dropped.
Results
Vegetation
The ten plots were dominated by herbaceous plants, characterized by Calamagrostis canadensis (Michx.) P.Beauv. and Chamerion angustifolium (L.) J. Holub, species present at all sites (Fig.
Summary of plant species occurrences. GBIF ID: GBIF (http://www.gbif.org/) taxon identifier. f: frequency of occurrence, the proportion of all samples in which each taxonomic unit was detected.
|
order |
family |
scientific name |
GBIF ID |
f |
|
Apiales |
Apiaceae |
Conioselinum chinense (L.) Britton, Sterns & Poggenb. |
0.1 |
|
|
Apiales |
Apiaceae |
Heracleum maximum Bartr. |
0.3 |
|
|
Asterales |
Asteraceae |
Achillea borealis Bong. |
0.3 |
|
|
Asterales |
Asteraceae |
Senecio triangularis Hook. |
0.1 |
|
|
Caryophyllales |
Caryophyllaceae |
Moehringia lateriflora (L.) Fenzl |
0.1 |
|
|
Dipsacales |
Adoxaceae |
Sambucus racemosa L. |
0.2 |
|
|
Equisetales |
Equisetaceae |
Equisetum arvense L. |
0.4 |
|
|
Equisetales |
Equisetaceae |
Equisetum L. |
0.5 |
|
|
Equisetales |
Equisetaceae |
Equisetum sylvaticum L. |
0.2 |
|
|
Ericales |
Ericaceae |
Pyrola asarifolia Michx. |
0.2 |
|
|
Ericales |
Ericaceae |
Vaccinium caespitosum Michaux |
0.1 |
|
|
Ericales |
Ericaceae |
Vaccinium vitis-idaea L. |
0.1 |
|
|
Ericales |
Polemoniaceae |
Polemonium acutiflorum Willd. ex Roem. & Schult. |
0.1 |
|
|
Ericales |
Primulaceae |
Trientalis europaea L. |
0.1 |
|
|
Fabales |
Fabaceae |
Lupinus nootkatensis Sims |
0.6 |
|
|
Fagales |
Betulaceae |
Alnus Mill. |
0.1 |
|
|
Gentianales |
Gentianaceae |
Swertia perennis L. |
0.2 |
|
|
Gentianales |
Rubiaceae |
Galium L. |
0.2 |
|
|
Geraniales |
Geraniaceae |
Geranium erianthum DC. |
0.6 |
|
|
Lamiales |
Orobanchaceae |
Castilleja unalaschcensis (Cham. & Schltdl.) Malte |
0.4 |
|
|
Liliales |
Liliaceae |
Streptopus amplexifolius (L.) DC. |
0.9 |
|
|
Liliales |
Melanthiaceae |
Veratrum viride Aiton |
0.7 |
|
|
Malpighiales |
Salicaceae |
Populus tremuloides Michx. |
0.1 |
|
|
Malpighiales |
Salicaceae |
Salix barclayi Anderss. |
0.1 |
|
|
Malpighiales |
Salicaceae |
Salix L. |
0.2 |
|
|
Malpighiales |
Violaceae |
Viola L. |
0.1 |
|
|
Myrtales |
Onagraceae |
Chamerion angustifolium (L.) J. Holub |
1.0 |
|
|
Pinales |
Pinaceae |
Picea lutzii Little |
0.1 |
|
|
Poales |
Cyperaceae |
Carex macrochaeta C.A.Mey. |
0.1 |
|
|
Poales |
Cyperaceae |
Carex mertensii J.D.Prescott ex Bong. |
0.2 |
|
|
Poales |
Juncaceae |
Luzula parviflora (Ehrh.) Desv. |
0.2 |
|
|
Poales |
Poaceae |
Alopecurus magellanicus Lam. |
0.1 |
|
|
Poales |
Poaceae |
Calamagrostis canadensis (Michx.) P.Beauv. |
1.0 |
|
|
Poales |
Poaceae |
Festuca altaica Trin. |
0.1 |
|
|
Poales |
Poaceae |
Phleum alpinum L. |
0.1 |
|
|
Poales |
Poaceae |
Poa arctica R.Br. |
0.1 |
|
|
Polypodiales |
Athyriaceae |
Athyrium filix-femina (L.) Roth |
0.5 |
|
|
Polypodiales |
Cystopteridaceae |
Gymnocarpium dryopteris (L.) Newm. |
0.1 |
|
|
Polypodiales |
Dryopteridaceae |
Dryopteris expansa (C. Presl) Fraser-Jenk. & Jermy |
0.6 |
|
|
Ranunculales |
Ranunculaceae |
Aconitum delphiniifolium Hort.Prag. ex Steud. |
0.4 |
|
|
Rosales |
Rosaceae |
Rubus arcticus L. |
0.1 |
|
|
Rosales |
Rosaceae |
Rubus idaeus L. |
0.1 |
|
|
Rosales |
Rosaceae |
Rubus L. |
0.1 |
|
|
Rosales |
Rosaceae |
Rubus pedatus Sm. |
0.2 |
|
|
Rosales |
Rosaceae |
Sanguisorba canadensis L. |
0.7 |
|
|
Rosales |
Rosaceae |
Spiraea stevenii (Schneid.) Rydb. |
0.2 |
Study site 73F (
Oligochaetes
At three sites (30% of sites) we detected a single earthworm species, Dendrobaena octaedra (Savigny, 1826). From another site a single specimen (Arctos GUID: KNWR:Ento:10822) was identified as an enchytraeid based on its COI sequence (BOLD Process ID: MOBIL1272-16). This sequence differed from all other sequences on BOLD, founding a new BIN (BOLD:ADC0663) with a nearest neighbor identified as Mesenchytraeus orcae Eisen, 1904 (pairwise-distance: 3.51%). Collection data for oligochaetes are provided in Suppl. material
Arthropod morphological identifications
Based on tallies of the sample contents by sight identifications, the sweep net samples contained 22–325 (mean=103, SE=26) individuals per sample, a total of 1,029 specimens (Table
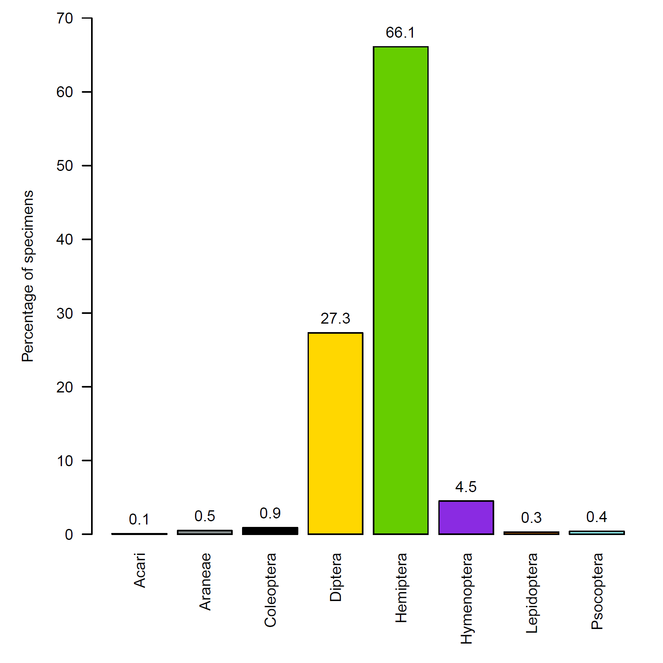
The samples were dominated by Hemiptera (66% of total specimens), especially the family Cicadellidae (25% of total specimens), and by Diptera (27% of total specimens). Hymenoptera represented only 4.5% of the specimens while Acari, Araneae, Coleoptera, Lepidoptera, and Psocoptera each represented less than 1% of specimens.
Arthropod metagenomic identifications
Sequencing yielded 30,672–54,228 reads per sample (mean=45,194, SD=7,601), a total of 451,941 reads. At the end of analysis and filtering steps, 391,316 identified reads were included in the occurrence data, 26,066–47,402 reads per sample (mean=39,132, SD=7,064) representing seven orders (Fig.
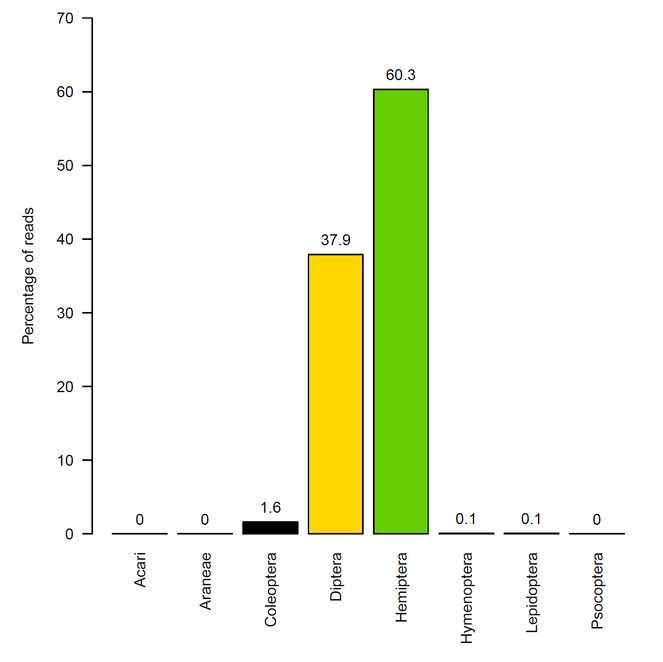
Of the 391,316 reads included in the occurrence data, these were dominated by Hemiptera (60%) and Diptera (38%). Coleoptera made up 1.6% of the reads while Araneae, Hymenoptera, Lepidoptera, and Psocoptera each included less than 1% of reads. No reads of Acari were identified.
Including provisional names, the metagenomic analysis yielded 67 unique taxon names (Table
Of the two species identifications we were able to make by sight, both were detected and identified by the metagenomic analysis. Misumena vatia (Clerck, 1757) was detected in the same sample in both the sight identifications and the metegomic data. Lauxania shewelli Perusse & Wheeler, 2000 was recorded at six sites in the sight identifications and detected at five of these same six sites in the metagenomic analysis.
Scrutiny of the remaining sequences that did not match anything in our reference database revealed a total of ten reads of human sequences from three sites.
Discussion
General characterization
Within the Calamagrostis-dominated grasslands of the Caribou Hills region we documented an entomofauna dominated by Hemiptera and Diptera, comparable to the general composition of sweep net samples collected in a Montana grassland by
We collected a similar number of specimens in ten 100 m2 sweep net samples over two days as
Although our arthropod sampling methods captured only a portion of the total arthropod fauna that would have been present, our results portrayed a reasonable snapshot of at least the fauna present on vegetation. All arthropods we documented are believed to be native to Alaska.
Comments on selected taxa
The single exotic earthworm species we collected, Dendrobaena octaedra, present at 30% of sites in our study area, was already known to be widespread on the Kenai Peninsula. This species was found at 70% of sites sampled on the Kenai National Wildlife Refuge, adjacent to our present study area, by
We assume that the single enchytraeid we collected was native because enchytraeids are widespread and diverse in southern Alaska (see
The chrysomelid beetle Altica tombacina was documented at two sites in the metagenomic analysis. Review of the notes associated with the specimen records on Arctos showed that these had been larvae when collected and so would have been unlikely to be identifiable based on morphology. Altica tombacina is to be expected in the study area, having been described from the Russian River vicinity (
The two staphylinid beetles seen in our samles were missed by our metagenomic methods likely due to their generally small size, primer bias, or a combination of these two reasons.
One of the more frequently detected species was Coenosia impunctata Malloch, 1920 (Diptera: Muscidae), found at seven sites. This species, described from Mount Katmai, Alaska (
Pipunculidae, specialist parasitoids on Cicadellidae and Delphacidae that are easly recognized at the family level, were seen in only two of the samples, but reads were detected in six samples in the metagenomic analysis, representing three species. At least some of these reads almost certainly came from pipunculid larvae within their cicadellid hosts.
Cicadellidae were well represented in our metagenomic data both in terms of read abundance and diversity, consistent with the high abundance and diversity of cicadellids documented from Canadian grasslands (
Of the Cicadellidae, the most common was Sonronius dahlbomi (Zetterstedt, 1840), detected at eight out of ten sites. According to
An entity bearing the provisional name of "Euscelis monodens sp. nov" (BIN BOLD:ACG7815) was the next most common cicadellid, detected at five sites. This provisional species is currently represented on BOLD by 15 specimens from British Columbia and the Yukon.
Delphacidae, herbivores of graminoids previously found by
It was noteworthy that Nabis (Nabidae) specimens were seen in the samples at four sites, but these were not detected by the metagenomic analysis despite these being some of the largest specimens in the samples, representing a significant portion of the material by body mass.
Irbisia sericans (Stål, 1858) (Hemiptera: Miridae), which we detected at one site, had previously been documented from Calamagrostis-dominiated grassland on the southern Kenai Peninsula where they had caused chlorosis of Calamagrostis leaves and stunting of the plants (
Human COI sequences in our data may have been due to contamination in our processing steps, but these may alternatively have come from human blood within biting flies collected in our samples. Biting flies (Simulium or Symphoromyia) were detected in all three samples where human sequences were detected (see Suppl. material
Metabarcoding as an identification method
The overall metagenomic results were consistent with our accounting of the specimens by eye, consistently portraying a community dominated by Hemiptera and Diptera. Our metagenomic methods under-represented the Araneae, Hemiptera, Hymenoptera, Lepidoptera, and Psocoptera while over-representing Coleoptera and Diptera relative to the proportions of specimens, likely due to primer bias during the PCR step. This is consistent with the experience of
To date, the purpose of most studies of involving HTS metabarcoding of arthropods has generally been to test and refine these methods (see
Our metabarcoding methods yielded timely (about 80 days including lab processing, shipping time, and analysis steps) and relatively inexpensive identifications ($US 1,115 for 131 sample × taxon identifications, $US 8.51 per identification). This is considerably more expensive than the < $US 0.40 chemical cost per identification of
There is an obvious trade-off between curating individual specimens for long-term deposition in an institutional repository and homogenizing specimens for HTS. Archiving individual specimens would have the potential to yield the most information as the specimens can be photographed, identified, and sequenced individually, and the specimens remain available for use in subsequent work. Rare and small species, easily missed by our HTS metagenomic methods, would be more likely to be detected using specimen-based, morphological methods.
However, processing and identification of thousands of specimens is time-consuming (
A non-destructive metabarcoding method (
Conclusions
We documented a native grassland arthropod fauna dominated by Hemiptera and Diptera. We found a single, epigeic, exotic earthworm species, but earthworms are unlikely to significantly alter these grassland communities unless additional exotic earthworms become established. We also demonstrated the usefulness of high-throughput sequencing metabarcoding as a tool for bioassessment of terrestrial arthropod assemblages.
Acknowledgements
We thank the Alaska Department of Natural Resources and Cook Inlet Region, Inc. for permission to collect samples on their lands. Derek S. Sikes (University of Alaska Museum, Fairbanks, Alaska), Bruce A. Snyder (Georgia College, Milledgeville, Georgia), and an anonymous reviewer provided comments that substantially improved this manuscript.
Author contributions
This project was conceived and planned by JMM, DM, and MB. Field work was carried out by JMM, DM, MB, and MO. MB performed initial sorting of samples, JDM was responsible for metagemonic processing, and MB conducted the final metagemonic analysis. The manuscript was prepared by MB.
References
- Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment.Frontiers in Marine Science3:96. https://doi.org/10.3389/fmars.2016.00096
- Biomonitoring 2.0: a new paradigm in ecosystem assessment made possible by next-generation DNA sequencing.Molecular Ecology21(8):2039‑2044. https://doi.org/10.1111/j.1365-294x.2012.05519.x
- Leafhoppers (Homoptera: Cicadellidae) of Canada and Alaska.Memoirs of the Entomological Society of Canada88:1‑180. https://doi.org/10.4039/entm8802fv
- Spruce beetle outbreaks on the Kenai Peninsula, Alaska, and Kluane National Park and Reserve, Yukon Territory: relationship to summer temperatures and regional differences in disturbance regimes.Forest Ecology and Management227(3):219‑232. https://doi.org/10.1016/j.foreco.2006.02.038
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: A web-based genome analysis tool for experimentalists. Current Protocols in Molecular Biology. https://doi.org/10.1002/0471142727.mb1910s89
- Manipulation of FASTQ data with Galaxy.Bioinformatics26(14):1783‑1785. https://doi.org/10.1093/bioinformatics/btq281
- Defining operational taxonomic units using DNA barcode data.Philosophical Transactions of the Royal Society B: Biological Sciences360(1462):1935‑1943. https://doi.org/10.1098/rstb.2005.1725
- White spruce regeneration following a major spruce beetle outbreak in forests on the Kenai Peninsula, Alaska.Forest Ecology and Management255(10):3571‑3579. https://doi.org/10.1016/j.foreco.2008.02.039
- Vegetation change and forest regeneration on the Kenai Peninsula, Alaska following a spruce beetle outbreak, 1987–2000.Forest Ecology and Management227(3):233‑246. https://doi.org/10.1016/j.foreco.2006.02.051
- Bowser M (2009) Modeling and Monitoring Terrestrial Arthropod Biodiversity on the Kenai National Wildlife Refuge, Alaska.University of Alaska Fairbanks,Fairbanks, Alaska,155pp. URL: http://www.fws.gov/uploadedFiles/Bowser_ML_2009.pdf
- DNA metabarcoding of insects and allies: an evaluation of primers and pipelines.Bulletin of Entomological Research105(6):717‑727. https://doi.org/10.1017/s0007485315000681
- A comparison of vacuum sampling versus sweep-netting for arthropod biodiversity measurements in California coastal sage scrub.Journal of Insect Conservation2(2):99‑106. https://doi.org/10.1023/a:1009653021706
- Human-facilitated invasion of exotic earthworms into northern boreal forests.Ecoscience14(4):482‑490. https://doi.org/10.2980/1195-6860(2007)14[482:hioeei]2.0.co;2
- Environmental monitoring using next generation sequencing: rapid identification of macroinvertebrate bioindicator species.Frontiers in Zoology10(1):45. https://doi.org/10.1186/1742-9994-10-45
- bold: Interface to Bold Systems 'API'.0.3.5. URL: http://CRAN.R-project.org/package=bold
- Targeted gene enrichment and high-throughput sequencing for environmental biomonitoring: a case study using freshwater macroinvertebrates.Molecular Ecology Resources16(5):1240‑1254. https://doi.org/10.1111/1755-0998.12488
- Comparison of two methods for sampling invertebrates: vacuum and sweep-net sampling.Journal of Field Ornithology82(1):60‑67. https://doi.org/10.1111/j.1557-9263.2010.00308.x
- Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—sequence relationships with an innovative metabarcoding protocol.PLoS ONE10(7):e0130324. https://doi.org/10.1371/journal.pone.0130324
- Earthworm invasion into previously earthworm-free temperate and boreal forests.Biological Invasions8(6):1235‑1245. https://doi.org/10.1007/s10530-006-9019-3
- Galaxy: a platform for interactive large-scale genome analysis.Genome Research15(10):1451‑1455. https://doi.org/10.1101/gr.4086505
- Large-scale biomonitoring of remote and threatened ecosystems via high-throughput sequencing.PLOS ONE10(10):e0138432. https://doi.org/10.1371/journal.pone.0138432
- Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences.Genome Biology11(8):R86. https://doi.org/10.1186/gb-2010-11-8-r86
- FASTX-Toolkit: FASTQ/A short-reads pre-processing tools.1.0.0.The Hannon laboratory, Cold Spring Harbor laboratory. URL: http://hannonlab.cshl.edu/fastx_toolkit/
- Assessing biodiversity of a freshwater benthic macroinvertebrate community through non-destructive environmental barcoding of DNA from preservative ethanol.BMC Ecology12(1):28. https://doi.org/10.1186/1472-6785-12-28
- Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos.PLoS ONE6(4):e17497. https://doi.org/10.1371/journal.pone.0017497
- Changes in hardwood forest understory plant communities in response to European earthworm invasions.Ecology87(7):1637‑1649. https://doi.org/10.1890/0012-9658(2006)87[1637:cihfup]2.0.co;2
- Effects of European Earthworm Invasion on Soil Characteristics in Northern Hardwood Forests of Minnesota, USA.Ecosystems8(8):911‑927. https://doi.org/10.1007/s10021-005-0066-x
- Hamilton KGA, Whitcomb R (2010) Leafhoppers (Homoptera: Cicadellidae): a major family adapted to grassland habitats. Arthropods of Canadian Grasslands (Volume 1): Ecology and Interactions in Grassland Habitats. https://doi.org/10.3752/9780968932148.ch8
- Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences270(1512):313‑321. https://doi.org/10.1098/rspb.2002.2218
- Exotic earthworm invasions in North America: ecological and policy implications.BioScience52(9):801. https://doi.org/10.1641/0006-3568(2002)052[0801:eeiina]2.0.co;2
- Effects of earthworm invasion on plant species richness in northern hardwood orests.Conservation Biology21(4):997‑1008. https://doi.org/10.1111/j.1523-1739.2007.00740.x
- Flora of Alaska and Neighboring Territories.Stanford University Press,Stanford, California,xxii + 1008pp. [ISBN9780804706438]
- eMODIS: a user-friendly data source, U.S. Geological Survey Open-File Report 2010–1055.U.S. Department of the Interior, U.S. Geological Survey,Reston, Virginia,10pp. URL: http://pubs.usgs.gov/of/2010/1055/
- Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding.Ecology Letters16(10):1245‑1257. https://doi.org/10.1111/ele.12162
- A late Pleistocene megafauna, Kenai Peninsula, southcentral Alaska.Alaska Journal of Anthropology13(1):57‑68. URL: https://www.alaskaanthropology.org/product/volume-131-2015-pdf/
- A test of the ‘hot’ mustard extraction method of sampling earthworms.Soil Biology and Biochemistry34(4):549‑552. https://doi.org/10.1016/s0038-0717(01)00211-5
- A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents.Frontiers in Zoology10(1):34. https://doi.org/10.1186/1742-9994-10-34
- Descriptions of new North American Anthomyiidae (Diptera).Transactions of the American Entomological Society46(2):133‑196. URL: http://www.biodiversitylibrary.org/part/70436
- Dritter nachtrag zur kaefer-fauna der Nord-Amerikanischen laender des Russichen Reiches.Bulletin de la Société Impériale des Naturalistes de Moscou26:95‑273.
- Terrestrial arthropod biodiversity: planning a study and recommended sampling techniques.Supplement to the Bulletin of the Entomological Society of Canada26(1):1‑33. URL: http://www.biology.ualberta.ca/bsc/briefs/brterrestrial.htm
- Observations of a grass bug on native bluejoint ranges.Agroborealis12(1):15‑18. URL: http://hdl.handle.net/11122/1675
- The DNA barcoding UAMU Project: Testing the insect identification power of DNA barcoding technology.Newsletter of the Alaska Entomological Society8(1):14‑17. URL: http://www.akentsoc.org/doc/AKES_newsletter_2015_I.pdf
- $1 DNA barcodes for reconstructing complex phenomes and finding rare species in specimen-rich samples.Cladistics32(1):100‑110. https://doi.org/10.1111/cla.12115
- ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data.Bioinformatics25(19):2607‑2608. https://doi.org/10.1093/bioinformatics/btp450
- BOLD: the Barcode of Life Data System (http://www.barcodinglife.org).Molecular Ecology Notes7(3):355‑364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
- A DNA-based registry for all animal species: he Barcode Index Number (BIN) system.PLoS ONE8(7):e66213. https://doi.org/10.1371/journal.pone.0066213
- R: a language and environment for statistical computing.3.2.2.R Foundation for Statistical Computing. Release date:2015-8-14. URL: https://www.R-project.org/
- The earthworms (Lumbricidae and Sparganophilidae) of Ontario.Royal Ontario Museum,Toronto, Ontario,ix + 141pp. [ISBN0888541910] https://doi.org/10.5962/bhl.title.60740
- VSEARCH: a versatile open source tool for metagenomics.PeerJ4:e2584. https://doi.org/10.7717/peerj.2584
- Distribution and abundance of exotic earthworms within a boreal forest system in southcentral Alaska.NeoBiota28:67‑86. https://doi.org/10.3897/neobiota.28.5503
- Predicting Future Potential Climate-Biomes for the Yukon, Northwest Territories, and Alaska.University of Alaska Fairbanks,Fairbanks, Alaska,105pp. URL: https://www.snap.uaf.edu/sites/default/files/Cliomes-FINAL_0.pdf
- Comparison of sweep net., D-Vac., and absolute sampling., and diel variation of sweep net sampling estimates in lentils for pea aphid (Homoptera: Aphididae), nabids (Hemiptera: Nabidae), lady beetles (Coleoptera: Coccinellidae), and lacewings (Neuroptera: Chrysopidae).Journal of Economic Entomology82(2):491‑506. https://doi.org/10.1093/jee/82.2.491
- Climate variability and spruce beetle (Dendroctonus rufipennis) outbreaks in south-central and southwest Alaska.Ecology92(7):1459‑1470. https://doi.org/10.1890/10-1118.1
- Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens.Molecular Ecology Resources14(5):892‑901. https://doi.org/10.1111/1755-0998.12236
- Environmental DNA barcode sequence capture: targeted, PCR-free sequence capture for biodiversity analysis from bulk environmental samples.bioRxivn/ahttps://doi.org/10.1101/087437
- Shorthouse J, Larson D (2010) Introduction to the grasslands and grassland arthropods of Canada. In: Shorthouse JD, Floate KD (Eds) Arthropods of Canadian Grasslands (Volume 1): Ecology and Interactions in Grassland Habitats.Biological Survey of Canada,1-24pp. https://doi.org/10.3752/9780968932148.ch1
- Building a DNA barcode library of Alaska’s nonmarine arthropods.GenomeNAhttps://doi.org/10.1139/gen-2015-0203
- A Field Guide to Alaska Grasses.Education Resources Publishing,Cumming, Georgia,xix + 384pp. [ISBN9780615648866]
- Sweeping beauty: is grassland arthropod community composition effectively estimated by sweep netting?Ecology and Evolution3(10):3347‑3358. https://doi.org/10.1002/ece3.688
- Wetland Sedges of Alaska.Alaska Natural Heritage Program, Environmental and Natural Resources Institute, University of Alaska Anchorage,Anchorage, Alaska,138pp. URL: http://accs.uaa.alaska.edu/files/botany/publications/2003/Wetland_Sedges_Alaska.pdf
- Arthropod-grass communities: comparison of communities in Ohio and Alaska.Journal of Biogeography8(1):53. https://doi.org/10.2307/2844592
- Timm T (1999) Distribution of freshwater oligochaetes in the west and east coastal regions of the North Pacific Ocean. In: Healy BM, Reynoldson TB, Coates KA (Eds) Aquatic Oligochaetes: Proceedings of the 7th International Symposium on Aquatic Oligochaetes held in Presque Isle, Maine, USA, 18–22 August 1997.Springer Netherlands,Dordrecht. https://doi.org/10.1007/978-94-011-4207-6_7
- The Alaska Vegetation Classification.U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station,Portland, Oregon,278pp. URL: http://www.fs.fed.us/pnw/publications/pnw_gtr286/
- Anderson's Flora of Alaska.Brigham Young University Press,Provo, Utah,xvi + 724pp. [ISBN9780842507059]
- PEAR: a fast and accurate Illumina Paired-End reAd mergeR.Bioinformatics30(5):614‑620. https://doi.org/10.1093/bioinformatics/btt593
- Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification.GigaScience2(1). https://doi.org/10.1186/2047-217x-2-4
Supplementary materials
Observation-based occurrences of vascular plant species. Dates are given in ISO 8601 format.
Earthworm specimen data. Dates are given in ISO 8601 format.
Arthropod specimen counts by sight identification and sample. Columns labeled KNWR:Ento:10838–KNWR:Ento:10847 are GUIDs of corresponding records on Arctos.
GUID: Arctos globally unique identifiers for the arthropod samples. Cluster label: illumina labels of centroid sequence clusters. Read count: cluster read counts. Process id: BOLD process IDs for matched database sequences. Similarity: similarity value from VSearch search. BIN: BOLD Barcode Index Numbers. Nucleotides: cluster centroid sequences.
Arthropod occurrence data from the metagenomic analysis expressed as read counts. BIN: BOLD Barcode Index Number. Columns labeled KNWR:Ento:10838–KNWR:Ento:10847 are GUIDs of corresponding specimen records on Arctos.